Bayesian Sampler Examples
Examples of running each sampler avaiable in 3ML.
Before, that, let’s discuss setting up configuration default sampler with default parameters. We can set in our configuration a default algorithm and default setup parameters for the samplers. This can ease fitting when we are doing exploratory data analysis.
With any of the samplers, you can pass keywords to access their setups. Read each pacakges documentation for more details.
[1]:
from threeML import *
import numpy as np
import dynesty
from jupyterthemes import jtplot
%matplotlib inline
jtplot.style(context="talk", fscale=1, ticks=True, grid=False)
silence_warnings()
set_threeML_style()
Welcome to JupyROOT 6.22/02
13:44:56 WARNING The naima package is not available. Models that depend on it will not be functions.py:51 available
WARNING The GSL library or the pygsl wrapper cannot be loaded. Models that depend on it functions.py:72 will not be available.
WARNING The ebltable package is not available. Models that depend on it will not be absorption.py:37 available
[2]:
threeML_config.bayesian.default_sampler
[2]:
<Sampler.emcee: 'emcee'>
[3]:
threeML_config.bayesian.emcee_setup
[3]:
{'n_burnin': None, 'n_iterations': 500, 'n_walkers': 50, 'seed': 5123}
If you simply run bayes_analysis.sample() the default sampler and its default parameters will be used.
Let’s make some data to fit.
[4]:
sin = Sin(K=1, f=0.1)
sin.phi.fix = True
sin.K.prior = Log_uniform_prior(lower_bound=0.5, upper_bound=1.5)
sin.f.prior = Uniform_prior(lower_bound=0, upper_bound=0.5)
model = Model(PointSource("demo", 0, 0, spectral_shape=sin))
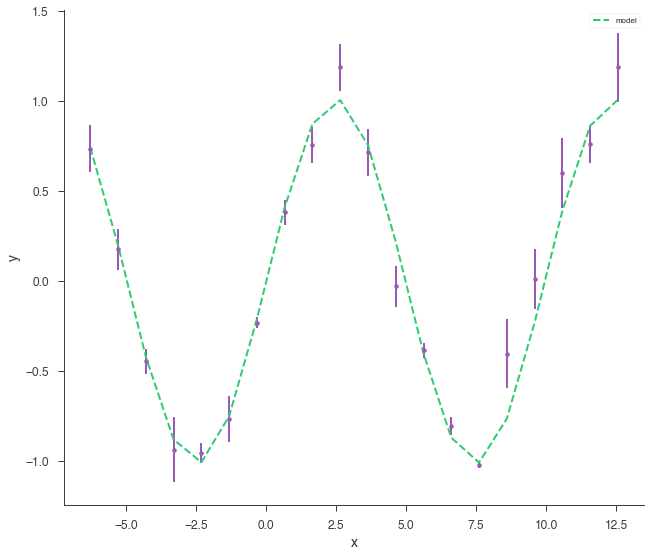
x = np.linspace(-2 * np.pi, 4 * np.pi, 20)
yerr = np.random.uniform(0.01, 0.2, 20)
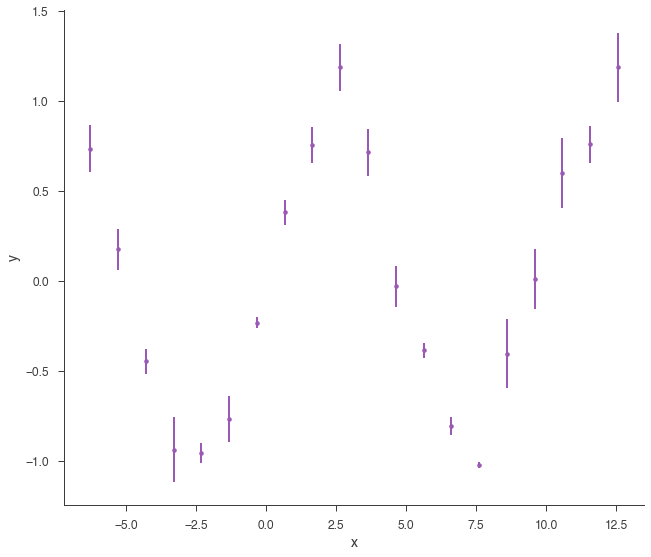
xyl = XYLike.from_function("demo", sin, x, yerr)
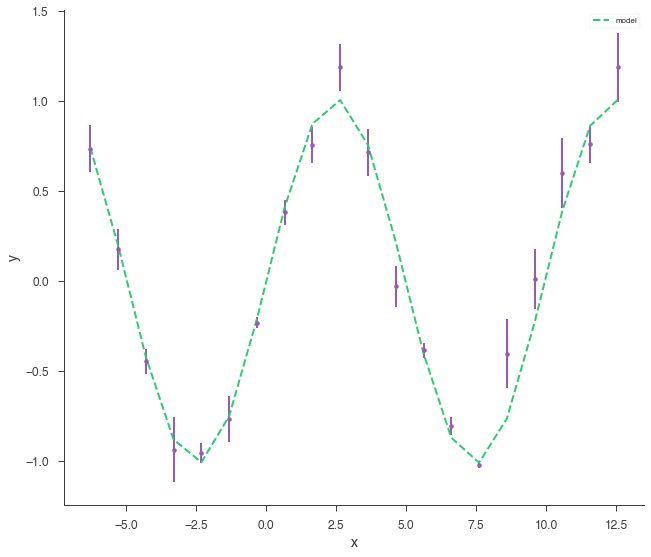
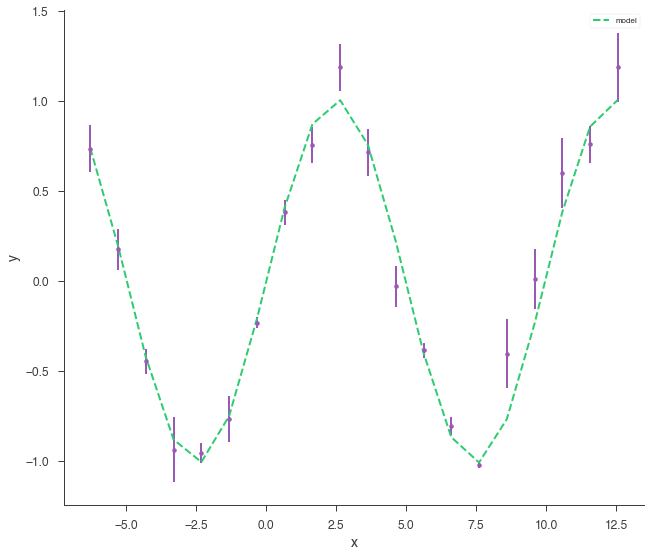
xyl.plot()
bayes_analysis = BayesianAnalysis(model, DataList(xyl))
emcee
[5]:
bayes_analysis.set_sampler("emcee")
bayes_analysis.sampler.setup(n_walkers=20, n_iterations=500)
bayes_analysis.sample()
xyl.plot()
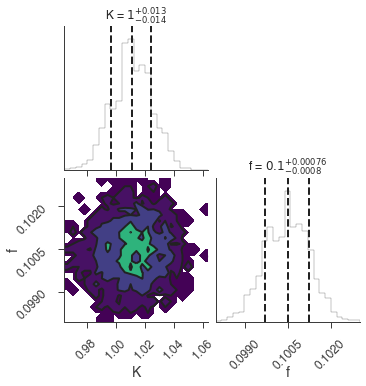
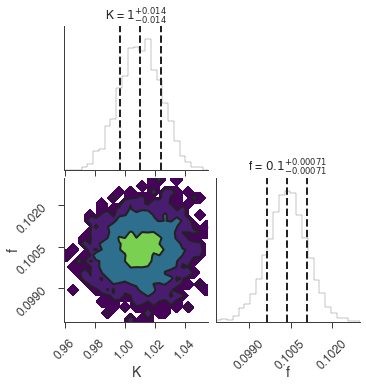
bayes_analysis.results.corner_plot()
13:45:01 INFO sampler set to emcee bayesian_analysis.py:233
13:45:05 INFO Mean acceptance fraction: 0.7174 emcee_sampler.py:157
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | 1.010 +/- 0.014 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (1.004 +/- 0.007) x 10^-1 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -10.507075 |
| total | -10.507075 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 25.720033 |
| BIC | 27.005615 |
| DIC | 24.908258 |
| PDIC | 1.942690 |
[5]:
multinest
[6]:
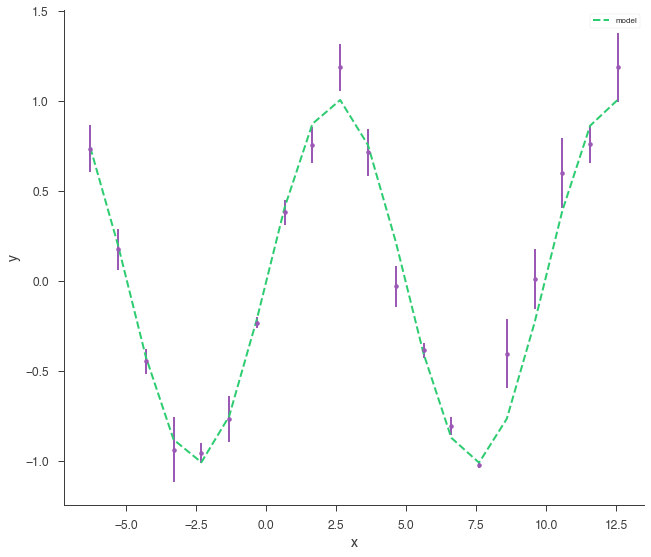
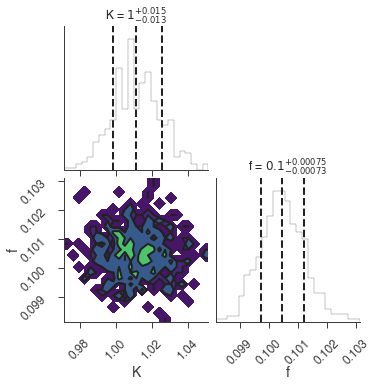
bayes_analysis.set_sampler("multinest")
bayes_analysis.sampler.setup(n_live_points=400, resume=False, auto_clean=True)
bayes_analysis.sample()
xyl.plot()
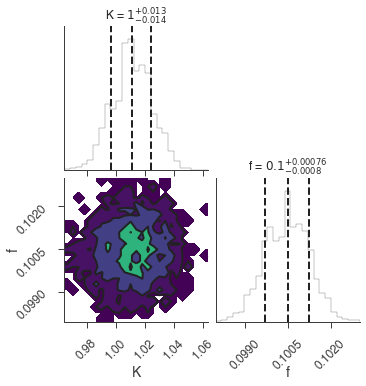
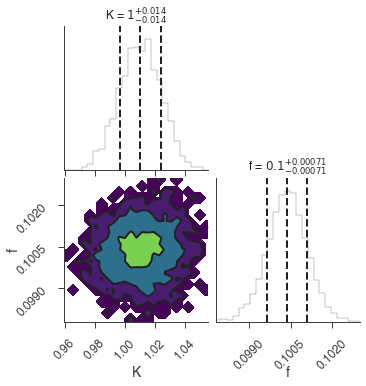
bayes_analysis.results.corner_plot()
13:45:08 INFO sampler set to multinest bayesian_analysis.py:233
analysing data from chains/fit-.txt
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | 1.012 -0.013 +0.014 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (1.005 -0.008 +0.007) x 10^-1 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -10.513789 |
| total | -10.513789 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 25.733460 |
| BIC | 27.019042 |
| DIC | 25.097662 |
| PDIC | 2.039027 |
| log(Z) | -8.462505 |
13:45:09 INFO deleting the chain directory chains multinest_sampler.py:261
WARNING:root:Too few points to create valid contours
[6]:
*****************************************************
MultiNest v3.10
Copyright Farhan Feroz & Mike Hobson
Release Jul 2015
no. of live points = 400
dimensionality = 2
*****************************************************
ln(ev)= -19.485638820181762 +/- 0.14121093259648074
Total Likelihood Evaluations: 5089
Sampling finished. Exiting MultiNest
dynesty
[7]:
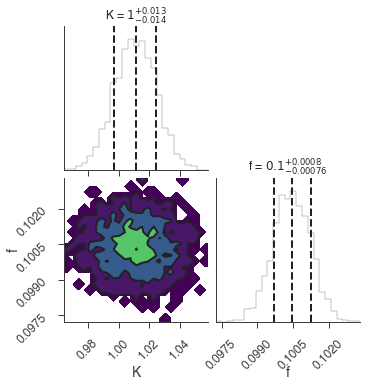
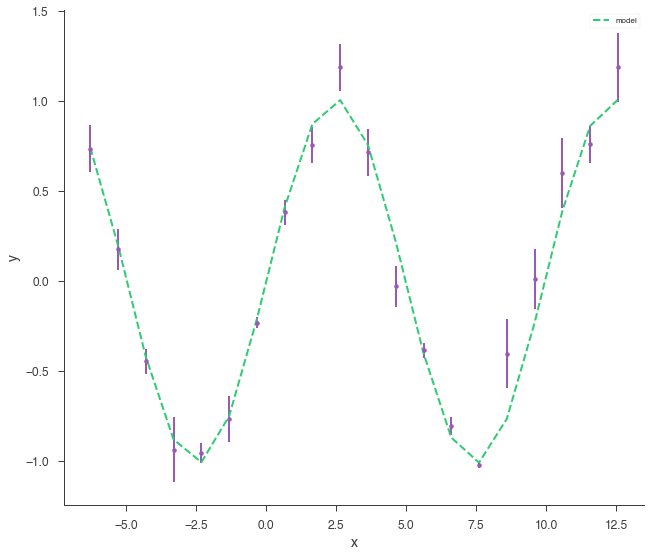
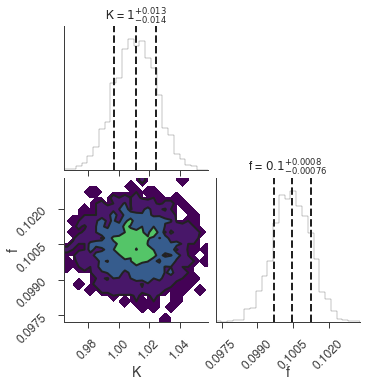
bayes_analysis.set_sampler("dynesty_nested")
bayes_analysis.sampler.setup(n_live_points=400)
bayes_analysis.sample()
xyl.plot()
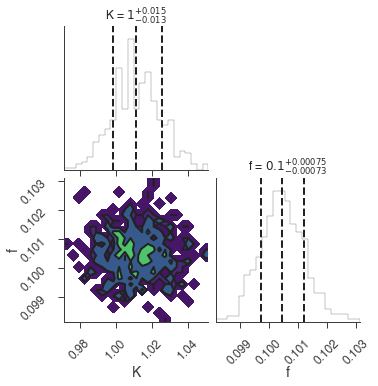
bayes_analysis.results.corner_plot()
13:45:10 INFO sampler set to dynesty_nested bayesian_analysis.py:233
3971it [00:10, 382.54it/s, +400 | bound: 8 | nc: 1 | ncall: 18560 | eff(%): 24.069 | loglstar: -inf < -10.498 < inf | logz: -19.376 +/- 0.141 | dlogz: 0.001 > 0.409]
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | 1.011 +/- 0.014 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (1.004 +/- 0.008) x 10^-1 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -10.509168 |
| total | -10.509168 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 25.724219 |
| BIC | 27.009801 |
| DIC | 25.166964 |
| PDIC | 2.076518 |
| log(Z) | -8.414852 |
[7]:
[8]:
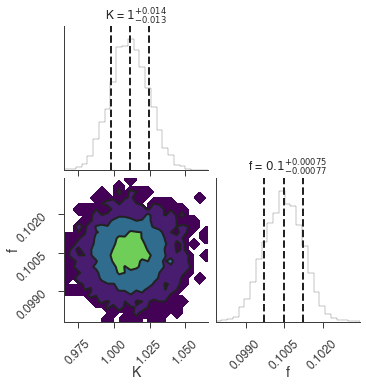
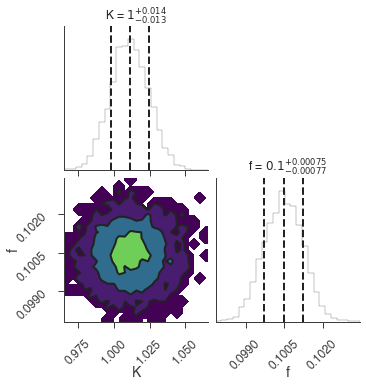
bayes_analysis.set_sampler("dynesty_dynamic")
bayes_analysis.sampler.setup(
stop_function=dynesty.utils.old_stopping_function, n_effective=None
)
bayes_analysis.sample()
xyl.plot()
bayes_analysis.results.corner_plot()
13:45:22 INFO sampler set to dynesty_dynamic bayesian_analysis.py:233
7229it [00:17, 1226.11it/s, batch: 0 | bound: 13 | nc: 1 | ncall: 26526 | eff(%): 27.223 | loglstar: -inf < -10.497 < inf | logz: -19.388 +/- 0.126 | dlogz: 0.001 > 0.010]
WARNING DeprecationWarning: This an old stopping function that will be removed in future releases
8344it [00:19, 1075.87it/s, batch: 1 | bound: 3 | nc: 1 | ncall: 28077 | eff(%): 29.692 | loglstar: -12.329 < -10.533 < -11.013 | logz: -19.388 +/- 0.130 | stop: 1.584]
WARNING DeprecationWarning: This an old stopping function that will be removed in future releases
8958it [00:21, 594.44it/s, batch: 2 | bound: 2 | nc: 1 | ncall: 28729 | eff(%): 31.156 | loglstar: -12.757 < -10.557 < -12.318 | logz: -19.380 +/- 0.106 | stop: 1.128]
WARNING DeprecationWarning: This an old stopping function that will be removed in future releases
9180it [00:22, 329.13it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 28976 | eff(%): 31.202 | loglstar: -13.169 < -12.298 < -12.752 | logz: -19.402 +/- 0.099 | stop: 1.005]
WARNING DeprecationWarning: This an old stopping function that will be removed in future releases
9570it [00:23, 408.19it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29421 | eff(%): 32.528 | loglstar: -13.169 < -10.500 < -12.752 | logz: -19.402 +/- 0.099 | stop: 0.984]
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | 1.011 -0.013 +0.014 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (1.005 +/- 0.008) x 10^-1 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -10.506859 |
| total | -10.506859 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 25.719601 |
| BIC | 27.005183 |
| DIC | 24.927015 |
| PDIC | 1.955806 |
| log(Z) | -8.433732 |
[8]:
zeus
[9]:
bayes_analysis.set_sampler("zeus")
bayes_analysis.sampler.setup(n_walkers=20, n_iterations=500)
bayes_analysis.sample()
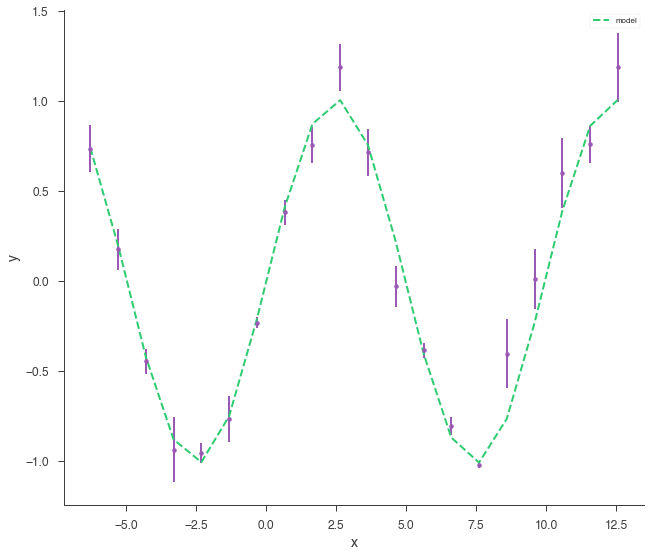
xyl.plot()
bayes_analysis.results.corner_plot()
13:45:48 INFO sampler set to zeus bayesian_analysis.py:233
WARNING:root:The sampler class has been deprecated. Please use the new EnsembleSampler class.
The run method has been deprecated and it will be removed. Please use the new run_mcmc method.
Initialising ensemble of 20 walkers...
Sampling progress : 100%|██████████| 625/625 [00:15<00:00, 39.75it/s]
Summary
-------
Number of Generations: 625
Number of Parameters: 2
Number of Walkers: 20
Number of Tuning Generations: 35
Scale Factor: 1.339496
Mean Integrated Autocorrelation Time: 3.38
Effective Sample Size: 3701.44
Number of Log Probability Evaluations: 65256
Effective Samples per Log Probability Evaluation: 0.056722
None
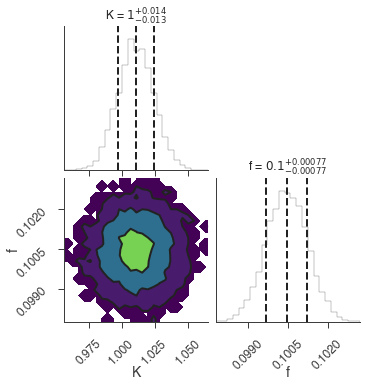
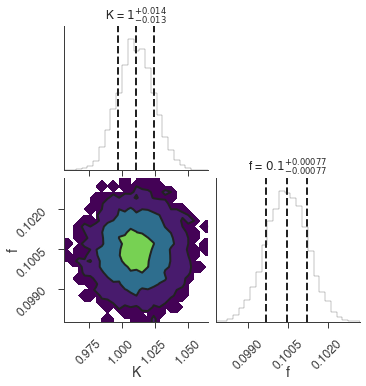
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | 1.011 +/- 0.014 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (1.005 +/- 0.008) x 10^-1 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -10.506904 |
| total | -10.506904 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 25.719690 |
| BIC | 27.005272 |
| DIC | 25.095113 |
| PDIC | 2.040639 |
[9]:
ultranest
[10]:
bayes_analysis.set_sampler("ultranest")
bayes_analysis.sampler.setup(
min_num_live_points=400, frac_remain=0.5, use_mlfriends=False
)
bayes_analysis.sample()
xyl.plot()
bayes_analysis.results.corner_plot()
13:46:05 INFO sampler set to ultranest bayesian_analysis.py:233
[ultranest] Sampling 400 live points from prior ...
[ultranest] Explored until L=-1e+01
[ultranest] Likelihood function evaluations: 8677
[ultranest] logZ = -19.37 +- 0.09592
[ultranest] Effective samples strategy satisfied (ESS = 974.7, need >400)
[ultranest] Posterior uncertainty strategy is satisfied (KL: 0.46+-0.05 nat, need <0.50 nat)
[ultranest] Evidency uncertainty strategy is satisfied (dlogz=0.42, need <0.5)
[ultranest] logZ error budget: single: 0.14 bs:0.10 tail:0.41 total:0.42 required:<0.50
[ultranest] done iterating.
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | 1.011 -0.014 +0.013 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (1.005 +/- 0.008) x 10^-1 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -10.507074 |
| total | -10.507074 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 25.720031 |
| BIC | 27.005613 |
| DIC | 25.100989 |
| PDIC | 2.042336 |
| log(Z) | -8.409488 |
[10]: