Bayesian Sampler Examples
Examples of running each sampler avaiable in 3ML.
Before, that, let’s discuss setting up configuration default sampler with default parameters. We can set in our configuration a default algorithm and default setup parameters for the samplers. This can ease fitting when we are doing exploratory data analysis.
With any of the samplers, you can pass keywords to access their setups. Read each pacakges documentation for more details.
[1]:
from threeML import *
from threeML.plugins.XYLike import XYLike
import numpy as np
import dynesty
from jupyterthemes import jtplot
%matplotlib inline
jtplot.style(context="talk", fscale=1, ticks=True, grid=False)
silence_warnings()
set_threeML_style()
22:18:38 WARNING The naima package is not available. Models that depend on it will not be functions.py:48 available
WARNING The GSL library or the pygsl wrapper cannot be loaded. Models that depend on it functions.py:69 will not be available.
22:18:39 WARNING The ebltable package is not available. Models that depend on it will not be absorption.py:33 available
[2]:
threeML_config.bayesian.default_sampler
[2]:
<Sampler.emcee: 'emcee'>
[3]:
threeML_config.bayesian.emcee_setup
[3]:
{'n_burnin': None, 'n_iterations': 500, 'n_walkers': 50, 'seed': 5123}
If you simply run bayes_analysis.sample() the default sampler and its default parameters will be used.
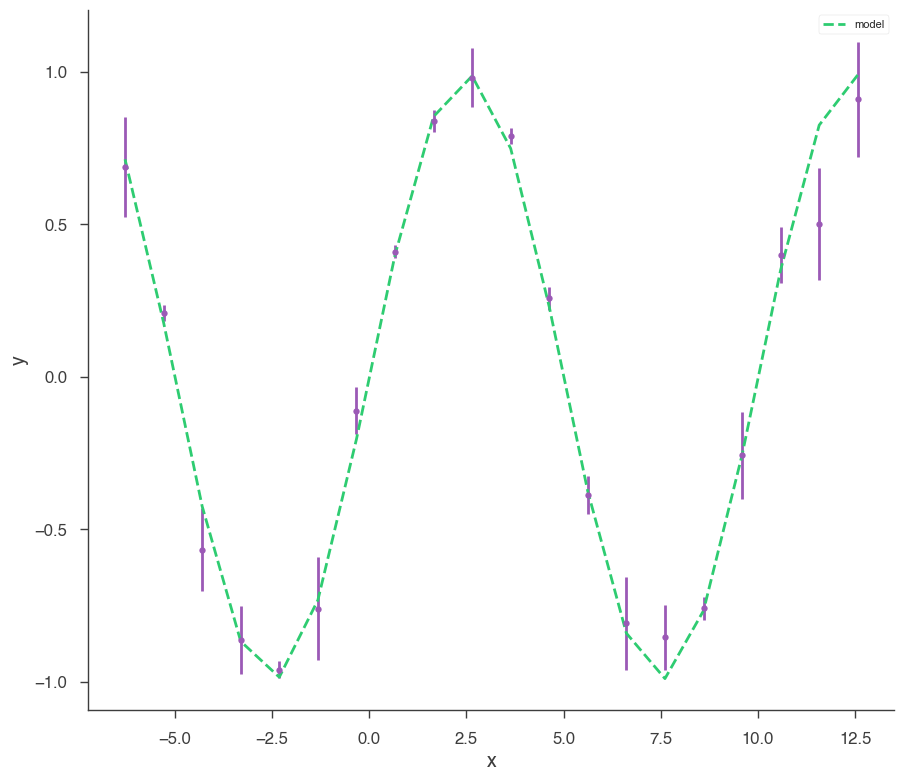
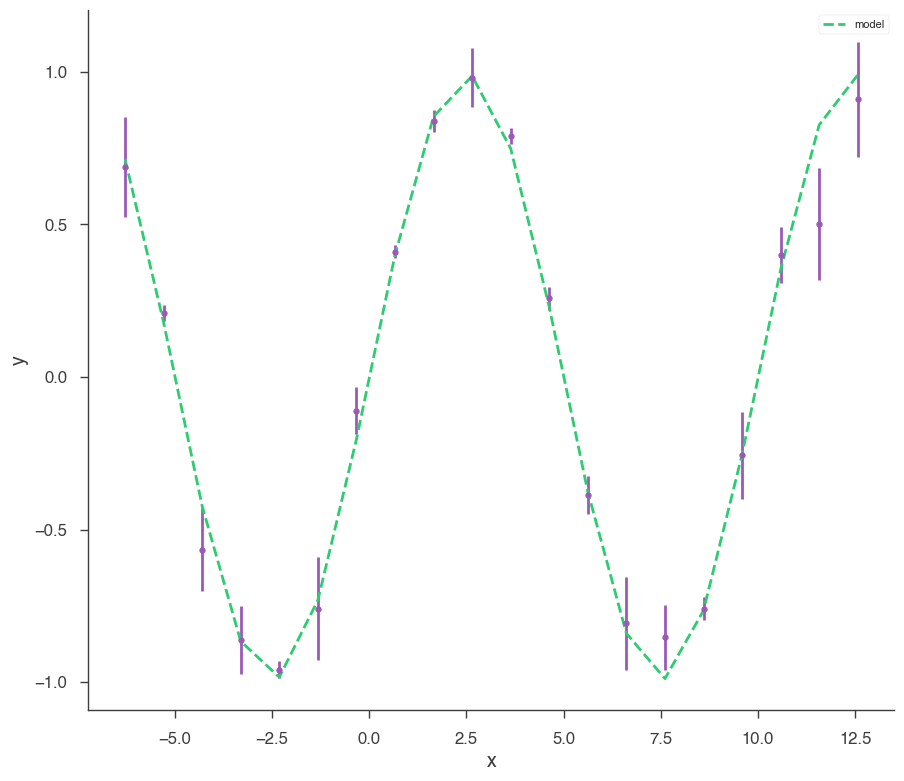
Let’s make some data to fit.
[4]:
sin = Sin(K=1, f=0.1)
sin.phi.fix = True
sin.K.prior = Log_uniform_prior(lower_bound=0.5, upper_bound=1.5)
sin.f.prior = Uniform_prior(lower_bound=0, upper_bound=0.5)
model = Model(PointSource("demo", 0, 0, spectral_shape=sin))
x = np.linspace(-2 * np.pi, 4 * np.pi, 20)
yerr = np.random.uniform(0.01, 0.2, 20)
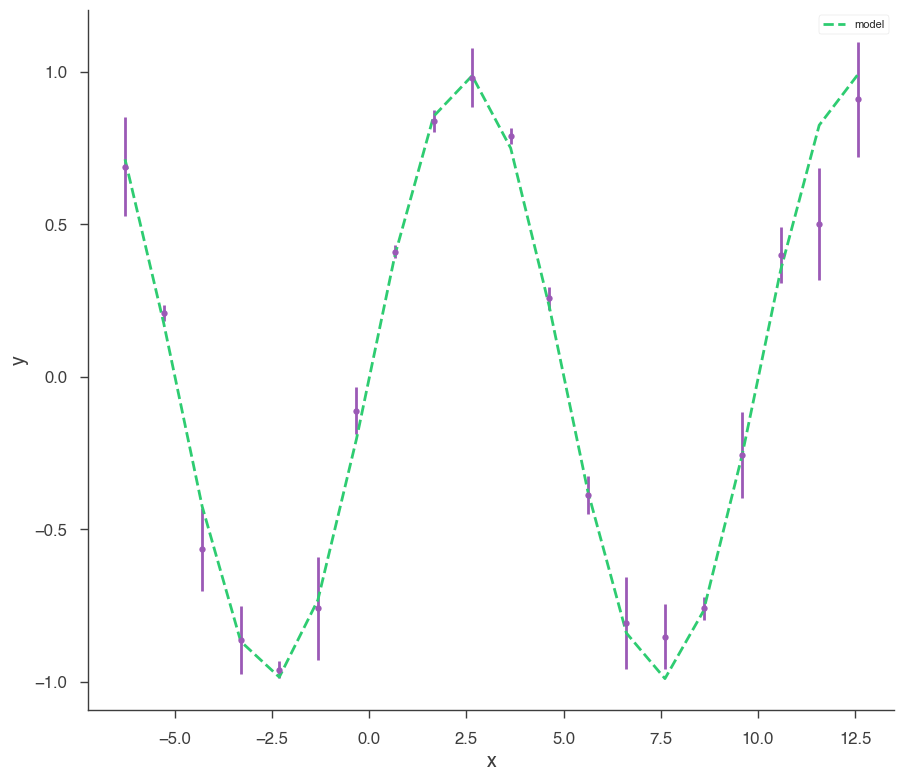
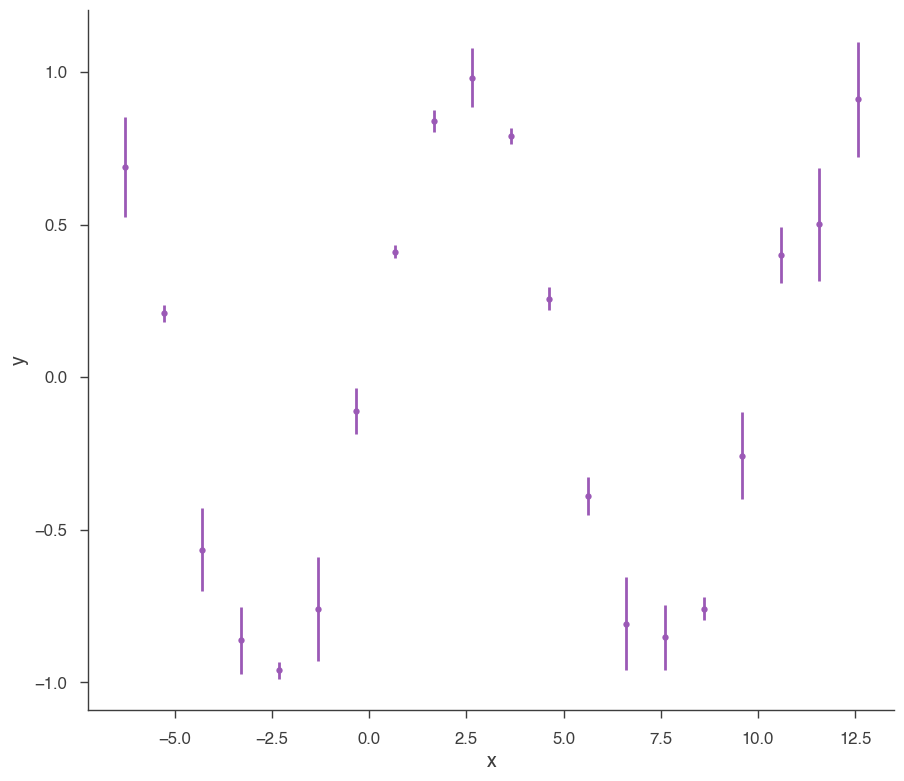
xyl = XYLike.from_function("demo", sin, x, yerr)
xyl.plot()
bayes_analysis = BayesianAnalysis(model, DataList(xyl))
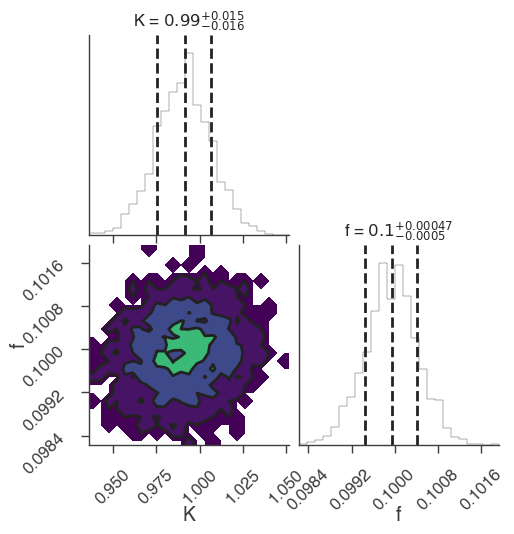
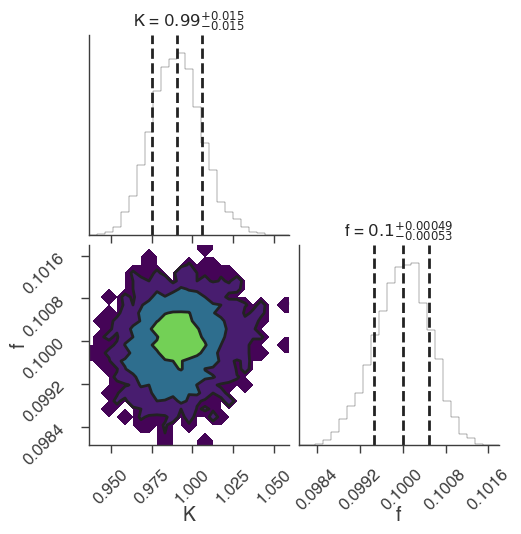
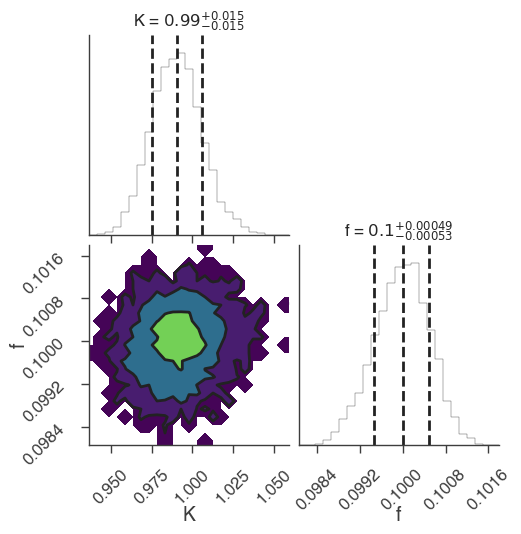
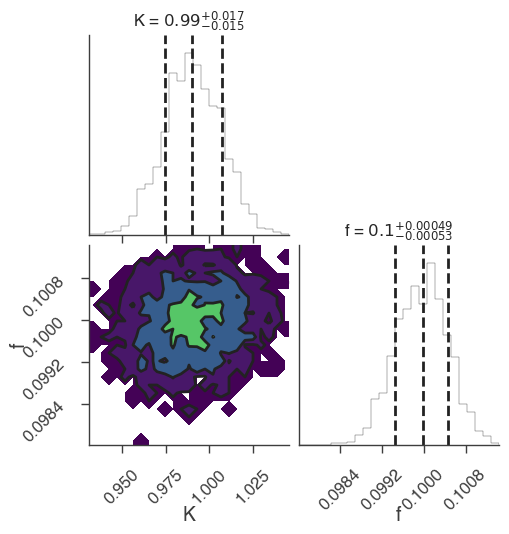
emcee
[5]:
bayes_analysis.set_sampler("emcee")
bayes_analysis.sampler.setup(n_walkers=20, n_iterations=500)
bayes_analysis.sample()
xyl.plot()
bayes_analysis.results.corner_plot()
INFO sampler set to emcee bayesian_analysis.py:202
22:18:46 INFO Mean acceptance fraction: 0.7144 emcee_sampler.py:157
22:18:47 INFO fit restored to maximum of posterior sampler_base.py:178
INFO fit restored to maximum of posterior sampler_base.py:178
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | (9.91 -0.16 +0.15) x 10^-1 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (9.99 -0.05 +0.06) x 10^-2 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -6.598494 |
| total | -6.598494 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 17.902870 |
| BIC | 19.188452 |
| DIC | 17.239380 |
| PDIC | 2.017942 |
[5]:
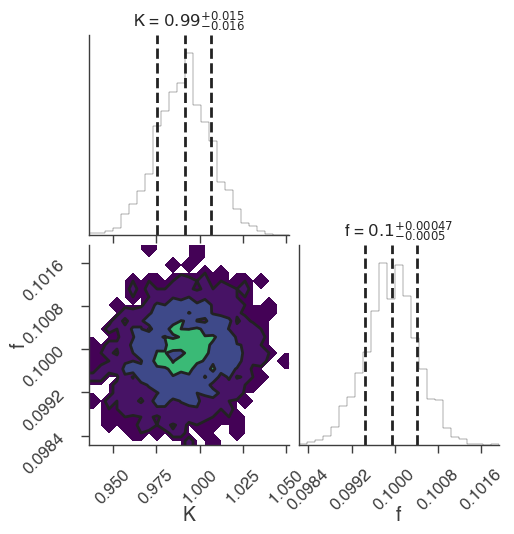
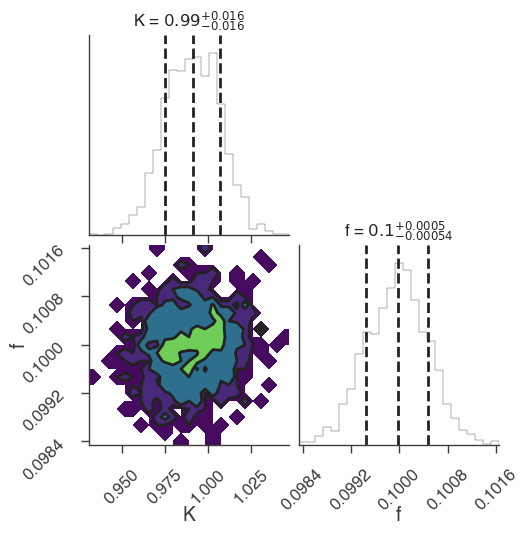
multinest
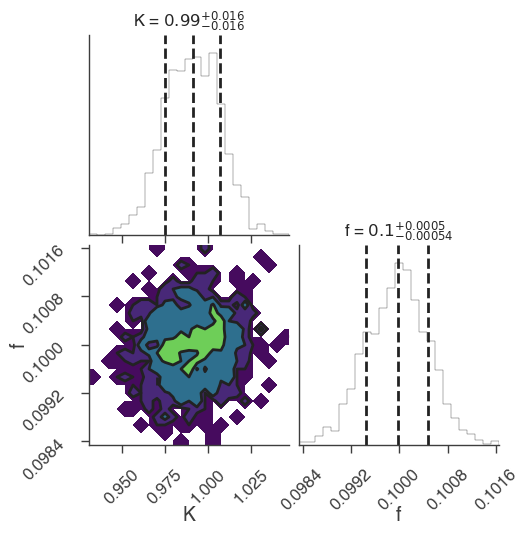
[6]:
bayes_analysis.set_sampler("multinest")
bayes_analysis.sampler.setup(n_live_points=400, resume=False, auto_clean=True)
bayes_analysis.sample()
xyl.plot()
bayes_analysis.results.corner_plot()
22:18:48 INFO sampler set to multinest bayesian_analysis.py:202
*****************************************************
MultiNest v3.10
Copyright Farhan Feroz & Mike Hobson
Release Jul 2015
no. of live points = 400
dimensionality = 2
*****************************************************
analysing data from chains/fit-.txt ln(ev)= -15.697095151737290 +/- 0.14206543454228004
Total Likelihood Evaluations: 6722
Sampling finished. Exiting MultiNest
22:18:50 INFO fit restored to maximum of posterior sampler_base.py:178
INFO fit restored to maximum of posterior sampler_base.py:178
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | (9.91 +/- 0.16) x 10^-1 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (10.00 +/- 0.05) x 10^-2 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -6.599193 |
| total | -6.599193 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 17.904268 |
| BIC | 19.189851 |
| DIC | 17.356304 |
| PDIC | 2.078925 |
| log(Z) | -6.817162 |
INFO deleting the chain directory chains multinest_sampler.py:255
[6]:
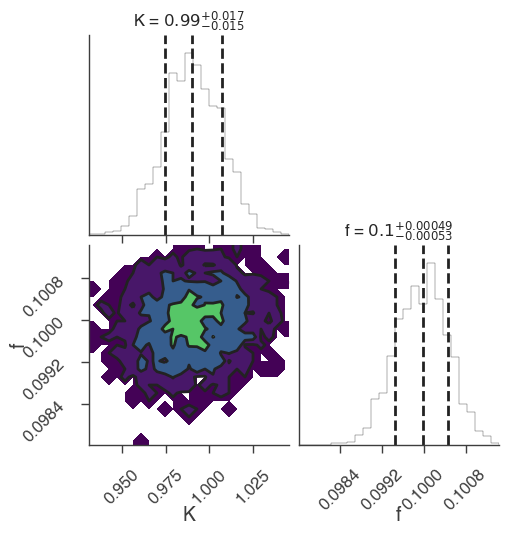
dynesty
[7]:
bayes_analysis.set_sampler("dynesty_nested")
bayes_analysis.sampler.setup(n_live_points=400)
bayes_analysis.sample()
xyl.plot()
bayes_analysis.results.corner_plot()
22:18:51 INFO sampler set to dynesty_nested bayesian_analysis.py:202
- 0it [00:00, ?it/s]
</pre>
- 0it [00:00, ?it/s]
end{sphinxVerbatim}
0it [00:00, ?it/s]
- 220it [00:00, 2199.80it/s, bound: 0 | nc: 1 | ncall: 694 | eff(%): 31.700 | loglstar: -inf < -3599.070 < inf | logz: -3606.304 +/- 0.134 | dlogz: 3606.224 > 0.409]
</pre>
- 220it [00:00, 2199.80it/s, bound: 0 | nc: 1 | ncall: 694 | eff(%): 31.700 | loglstar: -inf < -3599.070 < inf | logz: -3606.304 +/- 0.134 | dlogz: 3606.224 > 0.409]
end{sphinxVerbatim}
220it [00:00, 2199.80it/s, bound: 0 | nc: 1 | ncall: 694 | eff(%): 31.700 | loglstar: -inf < -3599.070 < inf | logz: -3606.304 +/- 0.134 | dlogz: 3606.224 > 0.409]
- 440it [00:00, 1427.83it/s, bound: 0 | nc: 1 | ncall: 1198 | eff(%): 36.728 | loglstar: -inf < -2641.566 < inf | logz: -2648.463 +/- 0.125 | dlogz: 2635.569 > 0.409]
</pre>
- 440it [00:00, 1427.83it/s, bound: 0 | nc: 1 | ncall: 1198 | eff(%): 36.728 | loglstar: -inf < -2641.566 < inf | logz: -2648.463 +/- 0.125 | dlogz: 2635.569 > 0.409]
end{sphinxVerbatim}
440it [00:00, 1427.83it/s, bound: 0 | nc: 1 | ncall: 1198 | eff(%): 36.728 | loglstar: -inf < -2641.566 < inf | logz: -2648.463 +/- 0.125 | dlogz: 2635.569 > 0.409]
- 599it [00:00, 1327.50it/s, bound: 0 | nc: 12 | ncall: 1765 | eff(%): 33.938 | loglstar: -inf < -2104.089 < inf | logz: -2111.894 +/- 0.133 | dlogz: 2099.304 > 0.409]
</pre>
- 599it [00:00, 1327.50it/s, bound: 0 | nc: 12 | ncall: 1765 | eff(%): 33.938 | loglstar: -inf < -2104.089 < inf | logz: -2111.894 +/- 0.133 | dlogz: 2099.304 > 0.409]
end{sphinxVerbatim}
599it [00:00, 1327.50it/s, bound: 0 | nc: 12 | ncall: 1765 | eff(%): 33.938 | loglstar: -inf < -2104.089 < inf | logz: -2111.894 +/- 0.133 | dlogz: 2099.304 > 0.409]
- 740it [00:00, 908.52it/s, bound: 0 | nc: 14 | ncall: 2626 | eff(%): 28.180 | loglstar: -inf < -1786.203 < inf | logz: -1794.122 +/- 0.136 | dlogz: 1781.018 > 0.409]
</pre>
- 740it [00:00, 908.52it/s, bound: 0 | nc: 14 | ncall: 2626 | eff(%): 28.180 | loglstar: -inf < -1786.203 < inf | logz: -1794.122 +/- 0.136 | dlogz: 1781.018 > 0.409]
end{sphinxVerbatim}
740it [00:00, 908.52it/s, bound: 0 | nc: 14 | ncall: 2626 | eff(%): 28.180 | loglstar: -inf < -1786.203 < inf | logz: -1794.122 +/- 0.136 | dlogz: 1781.018 > 0.409]
- 847it [00:00, 830.70it/s, bound: 0 | nc: 5 | ncall: 3458 | eff(%): 24.494 | loglstar: -inf < -1269.503 < inf | logz: -1277.256 +/- 0.134 | dlogz: 1263.202 > 0.409]
</pre>
- 847it [00:00, 830.70it/s, bound: 0 | nc: 5 | ncall: 3458 | eff(%): 24.494 | loglstar: -inf < -1269.503 < inf | logz: -1277.256 +/- 0.134 | dlogz: 1263.202 > 0.409]
end{sphinxVerbatim}
847it [00:00, 830.70it/s, bound: 0 | nc: 5 | ncall: 3458 | eff(%): 24.494 | loglstar: -inf < -1269.503 < inf | logz: -1277.256 +/- 0.134 | dlogz: 1263.202 > 0.409]
- 940it [00:01, 603.84it/s, bound: 0 | nc: 8 | ncall: 4371 | eff(%): 21.505 | loglstar: -inf < -953.279 < inf | logz: -962.180 +/- 0.146 | dlogz: 949.937 > 0.409]
</pre>
- 940it [00:01, 603.84it/s, bound: 0 | nc: 8 | ncall: 4371 | eff(%): 21.505 | loglstar: -inf < -953.279 < inf | logz: -962.180 +/- 0.146 | dlogz: 949.937 > 0.409]
end{sphinxVerbatim}
940it [00:01, 603.84it/s, bound: 0 | nc: 8 | ncall: 4371 | eff(%): 21.505 | loglstar: -inf < -953.279 < inf | logz: -962.180 +/- 0.146 | dlogz: 949.937 > 0.409]
- 1012it [00:01, 570.99it/s, bound: 0 | nc: 2 | ncall: 5196 | eff(%): 19.477 | loglstar: -inf < -783.982 < inf | logz: -792.386 +/- 0.141 | dlogz: 779.483 > 0.409]
</pre>
- 1012it [00:01, 570.99it/s, bound: 0 | nc: 2 | ncall: 5196 | eff(%): 19.477 | loglstar: -inf < -783.982 < inf | logz: -792.386 +/- 0.141 | dlogz: 779.483 > 0.409]
end{sphinxVerbatim}
1012it [00:01, 570.99it/s, bound: 0 | nc: 2 | ncall: 5196 | eff(%): 19.477 | loglstar: -inf < -783.982 < inf | logz: -792.386 +/- 0.141 | dlogz: 779.483 > 0.409]
- 1076it [00:01, 514.61it/s, bound: 0 | nc: 66 | ncall: 6231 | eff(%): 17.268 | loglstar: -inf < -635.304 < inf | logz: -644.275 +/- 0.142 | dlogz: 632.621 > 0.409]
</pre>
- 1076it [00:01, 514.61it/s, bound: 0 | nc: 66 | ncall: 6231 | eff(%): 17.268 | loglstar: -inf < -635.304 < inf | logz: -644.275 +/- 0.142 | dlogz: 632.621 > 0.409]
end{sphinxVerbatim}
1076it [00:01, 514.61it/s, bound: 0 | nc: 66 | ncall: 6231 | eff(%): 17.268 | loglstar: -inf < -635.304 < inf | logz: -644.275 +/- 0.142 | dlogz: 632.621 > 0.409]
- 1132it [00:01, 453.25it/s, bound: 0 | nc: 9 | ncall: 7073 | eff(%): 16.005 | loglstar: -inf < -528.944 < inf | logz: -537.234 +/- 0.141 | dlogz: 524.697 > 0.409]
</pre>
- 1132it [00:01, 453.25it/s, bound: 0 | nc: 9 | ncall: 7073 | eff(%): 16.005 | loglstar: -inf < -528.944 < inf | logz: -537.234 +/- 0.141 | dlogz: 524.697 > 0.409]
end{sphinxVerbatim}
1132it [00:01, 453.25it/s, bound: 0 | nc: 9 | ncall: 7073 | eff(%): 16.005 | loglstar: -inf < -528.944 < inf | logz: -537.234 +/- 0.141 | dlogz: 524.697 > 0.409]
- 1180it [00:01, 378.56it/s, bound: 0 | nc: 4 | ncall: 7936 | eff(%): 14.869 | loglstar: -inf < -468.288 < inf | logz: -476.661 +/- 0.140 | dlogz: 464.046 > 0.409]
</pre>
- 1180it [00:01, 378.56it/s, bound: 0 | nc: 4 | ncall: 7936 | eff(%): 14.869 | loglstar: -inf < -468.288 < inf | logz: -476.661 +/- 0.140 | dlogz: 464.046 > 0.409]
end{sphinxVerbatim}
1180it [00:01, 378.56it/s, bound: 0 | nc: 4 | ncall: 7936 | eff(%): 14.869 | loglstar: -inf < -468.288 < inf | logz: -476.661 +/- 0.140 | dlogz: 464.046 > 0.409]
- 1221it [00:02, 275.50it/s, bound: 0 | nc: 24 | ncall: 8654 | eff(%): 14.109 | loglstar: -inf < -433.610 < inf | logz: -441.790 +/- 0.136 | dlogz: 428.852 > 0.409]
</pre>
- 1221it [00:02, 275.50it/s, bound: 0 | nc: 24 | ncall: 8654 | eff(%): 14.109 | loglstar: -inf < -433.610 < inf | logz: -441.790 +/- 0.136 | dlogz: 428.852 > 0.409]
end{sphinxVerbatim}
1221it [00:02, 275.50it/s, bound: 0 | nc: 24 | ncall: 8654 | eff(%): 14.109 | loglstar: -inf < -433.610 < inf | logz: -441.790 +/- 0.136 | dlogz: 428.852 > 0.409]
- 1253it [00:02, 257.23it/s, bound: 0 | nc: 3 | ncall: 9284 | eff(%): 13.496 | loglstar: -inf < -402.224 < inf | logz: -411.203 +/- 0.142 | dlogz: 400.699 > 0.409]
</pre>
- 1253it [00:02, 257.23it/s, bound: 0 | nc: 3 | ncall: 9284 | eff(%): 13.496 | loglstar: -inf < -402.224 < inf | logz: -411.203 +/- 0.142 | dlogz: 400.699 > 0.409]
end{sphinxVerbatim}
1253it [00:02, 257.23it/s, bound: 0 | nc: 3 | ncall: 9284 | eff(%): 13.496 | loglstar: -inf < -402.224 < inf | logz: -411.203 +/- 0.142 | dlogz: 400.699 > 0.409]
- 1282it [00:02, 233.41it/s, bound: 0 | nc: 24 | ncall: 10065 | eff(%): 12.737 | loglstar: -inf < -375.649 < inf | logz: -384.053 +/- 0.140 | dlogz: 373.188 > 0.409]
</pre>
- 1282it [00:02, 233.41it/s, bound: 0 | nc: 24 | ncall: 10065 | eff(%): 12.737 | loglstar: -inf < -375.649 < inf | logz: -384.053 +/- 0.140 | dlogz: 373.188 > 0.409]
end{sphinxVerbatim}
1282it [00:02, 233.41it/s, bound: 0 | nc: 24 | ncall: 10065 | eff(%): 12.737 | loglstar: -inf < -375.649 < inf | logz: -384.053 +/- 0.140 | dlogz: 373.188 > 0.409]
- 1307it [00:02, 206.17it/s, bound: 0 | nc: 58 | ncall: 10904 | eff(%): 11.986 | loglstar: -inf < -354.543 < inf | logz: -362.949 +/- 0.139 | dlogz: 351.898 > 0.409]
</pre>
- 1307it [00:02, 206.17it/s, bound: 0 | nc: 58 | ncall: 10904 | eff(%): 11.986 | loglstar: -inf < -354.543 < inf | logz: -362.949 +/- 0.139 | dlogz: 351.898 > 0.409]
end{sphinxVerbatim}
1307it [00:02, 206.17it/s, bound: 0 | nc: 58 | ncall: 10904 | eff(%): 11.986 | loglstar: -inf < -354.543 < inf | logz: -362.949 +/- 0.139 | dlogz: 351.898 > 0.409]
- 1329it [00:02, 190.42it/s, bound: 0 | nc: 5 | ncall: 11531 | eff(%): 11.525 | loglstar: -inf < -335.383 < inf | logz: -344.730 +/- 0.146 | dlogz: 334.360 > 0.409]
</pre>
- 1329it [00:02, 190.42it/s, bound: 0 | nc: 5 | ncall: 11531 | eff(%): 11.525 | loglstar: -inf < -335.383 < inf | logz: -344.730 +/- 0.146 | dlogz: 334.360 > 0.409]
end{sphinxVerbatim}
1329it [00:02, 190.42it/s, bound: 0 | nc: 5 | ncall: 11531 | eff(%): 11.525 | loglstar: -inf < -335.383 < inf | logz: -344.730 +/- 0.146 | dlogz: 334.360 > 0.409]
- 1355it [00:02, 201.51it/s, bound: 0 | nc: 66 | ncall: 12204 | eff(%): 11.103 | loglstar: -inf < -317.472 < inf | logz: -325.786 +/- 0.135 | dlogz: 314.479 > 0.409]
</pre>
- 1355it [00:02, 201.51it/s, bound: 0 | nc: 66 | ncall: 12204 | eff(%): 11.103 | loglstar: -inf < -317.472 < inf | logz: -325.786 +/- 0.135 | dlogz: 314.479 > 0.409]
end{sphinxVerbatim}
1355it [00:02, 201.51it/s, bound: 0 | nc: 66 | ncall: 12204 | eff(%): 11.103 | loglstar: -inf < -317.472 < inf | logz: -325.786 +/- 0.135 | dlogz: 314.479 > 0.409]
- 1376it [00:03, 175.16it/s, bound: 0 | nc: 7 | ncall: 12924 | eff(%): 10.647 | loglstar: -inf < -306.168 < inf | logz: -314.589 +/- 0.136 | dlogz: 303.288 > 0.409]
</pre>
- 1376it [00:03, 175.16it/s, bound: 0 | nc: 7 | ncall: 12924 | eff(%): 10.647 | loglstar: -inf < -306.168 < inf | logz: -314.589 +/- 0.136 | dlogz: 303.288 > 0.409]
end{sphinxVerbatim}
1376it [00:03, 175.16it/s, bound: 0 | nc: 7 | ncall: 12924 | eff(%): 10.647 | loglstar: -inf < -306.168 < inf | logz: -314.589 +/- 0.136 | dlogz: 303.288 > 0.409]
- 1395it [00:03, 159.82it/s, bound: 0 | nc: 23 | ncall: 13505 | eff(%): 10.330 | loglstar: -inf < -292.596 < inf | logz: -301.609 +/- 0.141 | dlogz: 290.452 > 0.409]
</pre>
- 1395it [00:03, 159.82it/s, bound: 0 | nc: 23 | ncall: 13505 | eff(%): 10.330 | loglstar: -inf < -292.596 < inf | logz: -301.609 +/- 0.141 | dlogz: 290.452 > 0.409]
end{sphinxVerbatim}
1395it [00:03, 159.82it/s, bound: 0 | nc: 23 | ncall: 13505 | eff(%): 10.330 | loglstar: -inf < -292.596 < inf | logz: -301.609 +/- 0.141 | dlogz: 290.452 > 0.409]
- 1412it [00:03, 154.37it/s, bound: 0 | nc: 81 | ncall: 14041 | eff(%): 10.056 | loglstar: -inf < -280.538 < inf | logz: -289.660 +/- 0.140 | dlogz: 278.415 > 0.409]
</pre>
- 1412it [00:03, 154.37it/s, bound: 0 | nc: 81 | ncall: 14041 | eff(%): 10.056 | loglstar: -inf < -280.538 < inf | logz: -289.660 +/- 0.140 | dlogz: 278.415 > 0.409]
end{sphinxVerbatim}
1412it [00:03, 154.37it/s, bound: 0 | nc: 81 | ncall: 14041 | eff(%): 10.056 | loglstar: -inf < -280.538 < inf | logz: -289.660 +/- 0.140 | dlogz: 278.415 > 0.409]
- 1428it [00:03, 127.70it/s, bound: 1 | nc: 1 | ncall: 14184 | eff(%): 10.068 | loglstar: -inf < -270.608 < inf | logz: -279.627 +/- 0.142 | dlogz: 268.334 > 0.409]
</pre>
- 1428it [00:03, 127.70it/s, bound: 1 | nc: 1 | ncall: 14184 | eff(%): 10.068 | loglstar: -inf < -270.608 < inf | logz: -279.627 +/- 0.142 | dlogz: 268.334 > 0.409]
end{sphinxVerbatim}
1428it [00:03, 127.70it/s, bound: 1 | nc: 1 | ncall: 14184 | eff(%): 10.068 | loglstar: -inf < -270.608 < inf | logz: -279.627 +/- 0.142 | dlogz: 268.334 > 0.409]
- 1625it [00:03, 508.98it/s, bound: 1 | nc: 5 | ncall: 14482 | eff(%): 11.221 | loglstar: -inf < -179.605 < inf | logz: -188.411 +/- 0.142 | dlogz: 177.959 > 0.409]
</pre>
- 1625it [00:03, 508.98it/s, bound: 1 | nc: 5 | ncall: 14482 | eff(%): 11.221 | loglstar: -inf < -179.605 < inf | logz: -188.411 +/- 0.142 | dlogz: 177.959 > 0.409]
end{sphinxVerbatim}
1625it [00:03, 508.98it/s, bound: 1 | nc: 5 | ncall: 14482 | eff(%): 11.221 | loglstar: -inf < -179.605 < inf | logz: -188.411 +/- 0.142 | dlogz: 177.959 > 0.409]
- 1751it [00:03, 502.19it/s, bound: 2 | nc: 1 | ncall: 14772 | eff(%): 11.854 | loglstar: -inf < -136.241 < inf | logz: -145.300 +/- 0.143 | dlogz: 134.498 > 0.409]
</pre>
- 1751it [00:03, 502.19it/s, bound: 2 | nc: 1 | ncall: 14772 | eff(%): 11.854 | loglstar: -inf < -136.241 < inf | logz: -145.300 +/- 0.143 | dlogz: 134.498 > 0.409]
end{sphinxVerbatim}
1751it [00:03, 502.19it/s, bound: 2 | nc: 1 | ncall: 14772 | eff(%): 11.854 | loglstar: -inf < -136.241 < inf | logz: -145.300 +/- 0.143 | dlogz: 134.498 > 0.409]
- 1948it [00:04, 785.74it/s, bound: 2 | nc: 5 | ncall: 15058 | eff(%): 12.937 | loglstar: -inf < -84.709 < inf | logz: -94.115 +/- 0.144 | dlogz: 82.775 > 0.409]
</pre>
- 1948it [00:04, 785.74it/s, bound: 2 | nc: 5 | ncall: 15058 | eff(%): 12.937 | loglstar: -inf < -84.709 < inf | logz: -94.115 +/- 0.144 | dlogz: 82.775 > 0.409]
end{sphinxVerbatim}
1948it [00:04, 785.74it/s, bound: 2 | nc: 5 | ncall: 15058 | eff(%): 12.937 | loglstar: -inf < -84.709 < inf | logz: -94.115 +/- 0.144 | dlogz: 82.775 > 0.409]
- 2051it [00:04, 828.01it/s, bound: 2 | nc: 1 | ncall: 15290 | eff(%): 13.414 | loglstar: -inf < -67.147 < inf | logz: -76.206 +/- 0.142 | dlogz: 64.544 > 0.409]
</pre>
- 2051it [00:04, 828.01it/s, bound: 2 | nc: 1 | ncall: 15290 | eff(%): 13.414 | loglstar: -inf < -67.147 < inf | logz: -76.206 +/- 0.142 | dlogz: 64.544 > 0.409]
end{sphinxVerbatim}
2051it [00:04, 828.01it/s, bound: 2 | nc: 1 | ncall: 15290 | eff(%): 13.414 | loglstar: -inf < -67.147 < inf | logz: -76.206 +/- 0.142 | dlogz: 64.544 > 0.409]
- 2152it [00:04, 702.35it/s, bound: 3 | nc: 1 | ncall: 15455 | eff(%): 13.924 | loglstar: -inf < -54.108 < inf | logz: -63.644 +/- 0.146 | dlogz: 51.754 > 0.409]
</pre>
- 2152it [00:04, 702.35it/s, bound: 3 | nc: 1 | ncall: 15455 | eff(%): 13.924 | loglstar: -inf < -54.108 < inf | logz: -63.644 +/- 0.146 | dlogz: 51.754 > 0.409]
end{sphinxVerbatim}
2152it [00:04, 702.35it/s, bound: 3 | nc: 1 | ncall: 15455 | eff(%): 13.924 | loglstar: -inf < -54.108 < inf | logz: -63.644 +/- 0.146 | dlogz: 51.754 > 0.409]
- 2329it [00:04, 928.96it/s, bound: 3 | nc: 1 | ncall: 15739 | eff(%): 14.798 | loglstar: -inf < -36.758 < inf | logz: -45.985 +/- 0.145 | dlogz: 33.631 > 0.409]
</pre>
- 2329it [00:04, 928.96it/s, bound: 3 | nc: 1 | ncall: 15739 | eff(%): 14.798 | loglstar: -inf < -36.758 < inf | logz: -45.985 +/- 0.145 | dlogz: 33.631 > 0.409]
end{sphinxVerbatim}
2329it [00:04, 928.96it/s, bound: 3 | nc: 1 | ncall: 15739 | eff(%): 14.798 | loglstar: -inf < -36.758 < inf | logz: -45.985 +/- 0.145 | dlogz: 33.631 > 0.409]
- 2442it [00:04, 732.74it/s, bound: 4 | nc: 1 | ncall: 15988 | eff(%): 15.274 | loglstar: -inf < -29.726 < inf | logz: -38.897 +/- 0.142 | dlogz: 26.239 > 0.409]
</pre>
- 2442it [00:04, 732.74it/s, bound: 4 | nc: 1 | ncall: 15988 | eff(%): 15.274 | loglstar: -inf < -29.726 < inf | logz: -38.897 +/- 0.142 | dlogz: 26.239 > 0.409]
end{sphinxVerbatim}
2442it [00:04, 732.74it/s, bound: 4 | nc: 1 | ncall: 15988 | eff(%): 15.274 | loglstar: -inf < -29.726 < inf | logz: -38.897 +/- 0.142 | dlogz: 26.239 > 0.409]
- 2597it [00:04, 894.10it/s, bound: 4 | nc: 1 | ncall: 16213 | eff(%): 16.018 | loglstar: -inf < -22.615 < inf | logz: -31.631 +/- 0.142 | dlogz: 18.561 > 0.409]
</pre>
- 2597it [00:04, 894.10it/s, bound: 4 | nc: 1 | ncall: 16213 | eff(%): 16.018 | loglstar: -inf < -22.615 < inf | logz: -31.631 +/- 0.142 | dlogz: 18.561 > 0.409]
end{sphinxVerbatim}
2597it [00:04, 894.10it/s, bound: 4 | nc: 1 | ncall: 16213 | eff(%): 16.018 | loglstar: -inf < -22.615 < inf | logz: -31.631 +/- 0.142 | dlogz: 18.561 > 0.409]
- 2709it [00:04, 938.35it/s, bound: 4 | nc: 2 | ncall: 16422 | eff(%): 16.496 | loglstar: -inf < -18.949 < inf | logz: -28.305 +/- 0.143 | dlogz: 14.958 > 0.409]
</pre>
- 2709it [00:04, 938.35it/s, bound: 4 | nc: 2 | ncall: 16422 | eff(%): 16.496 | loglstar: -inf < -18.949 < inf | logz: -28.305 +/- 0.143 | dlogz: 14.958 > 0.409]
end{sphinxVerbatim}
2709it [00:04, 938.35it/s, bound: 4 | nc: 2 | ncall: 16422 | eff(%): 16.496 | loglstar: -inf < -18.949 < inf | logz: -28.305 +/- 0.143 | dlogz: 14.958 > 0.409]
- 2820it [00:05, 807.50it/s, bound: 5 | nc: 1 | ncall: 16641 | eff(%): 16.946 | loglstar: -inf < -15.840 < inf | logz: -25.171 +/- 0.145 | dlogz: 11.539 > 0.409]
</pre>
- 2820it [00:05, 807.50it/s, bound: 5 | nc: 1 | ncall: 16641 | eff(%): 16.946 | loglstar: -inf < -15.840 < inf | logz: -25.171 +/- 0.145 | dlogz: 11.539 > 0.409]
end{sphinxVerbatim}
2820it [00:05, 807.50it/s, bound: 5 | nc: 1 | ncall: 16641 | eff(%): 16.946 | loglstar: -inf < -15.840 < inf | logz: -25.171 +/- 0.145 | dlogz: 11.539 > 0.409]
- 2915it [00:05, 673.57it/s, bound: 5 | nc: 1 | ncall: 16784 | eff(%): 17.368 | loglstar: -inf < -14.080 < inf | logz: -23.229 +/- 0.144 | dlogz: 9.351 > 0.409]
</pre>
- 2915it [00:05, 673.57it/s, bound: 5 | nc: 1 | ncall: 16784 | eff(%): 17.368 | loglstar: -inf < -14.080 < inf | logz: -23.229 +/- 0.144 | dlogz: 9.351 > 0.409]
end{sphinxVerbatim}
2915it [00:05, 673.57it/s, bound: 5 | nc: 1 | ncall: 16784 | eff(%): 17.368 | loglstar: -inf < -14.080 < inf | logz: -23.229 +/- 0.144 | dlogz: 9.351 > 0.409]
- 3074it [00:05, 856.98it/s, bound: 5 | nc: 6 | ncall: 17094 | eff(%): 17.983 | loglstar: -inf < -11.544 < inf | logz: -20.805 +/- 0.144 | dlogz: 6.527 > 0.409]
</pre>
- 3074it [00:05, 856.98it/s, bound: 5 | nc: 6 | ncall: 17094 | eff(%): 17.983 | loglstar: -inf < -11.544 < inf | logz: -20.805 +/- 0.144 | dlogz: 6.527 > 0.409]
end{sphinxVerbatim}
3074it [00:05, 856.98it/s, bound: 5 | nc: 6 | ncall: 17094 | eff(%): 17.983 | loglstar: -inf < -11.544 < inf | logz: -20.805 +/- 0.144 | dlogz: 6.527 > 0.409]
- 3178it [00:05, 610.22it/s, bound: 6 | nc: 3 | ncall: 17258 | eff(%): 18.415 | loglstar: -inf < -10.500 < inf | logz: -19.751 +/- 0.143 | dlogz: 5.215 > 0.409]
</pre>
- 3178it [00:05, 610.22it/s, bound: 6 | nc: 3 | ncall: 17258 | eff(%): 18.415 | loglstar: -inf < -10.500 < inf | logz: -19.751 +/- 0.143 | dlogz: 5.215 > 0.409]
end{sphinxVerbatim}
3178it [00:05, 610.22it/s, bound: 6 | nc: 3 | ncall: 17258 | eff(%): 18.415 | loglstar: -inf < -10.500 < inf | logz: -19.751 +/- 0.143 | dlogz: 5.215 > 0.409]
- 3315it [00:05, 745.11it/s, bound: 6 | nc: 3 | ncall: 17474 | eff(%): 18.971 | loglstar: -inf < -9.407 < inf | logz: -18.668 +/- 0.144 | dlogz: 3.814 > 0.409]
</pre>
- 3315it [00:05, 745.11it/s, bound: 6 | nc: 3 | ncall: 17474 | eff(%): 18.971 | loglstar: -inf < -9.407 < inf | logz: -18.668 +/- 0.144 | dlogz: 3.814 > 0.409]
end{sphinxVerbatim}
3315it [00:05, 745.11it/s, bound: 6 | nc: 3 | ncall: 17474 | eff(%): 18.971 | loglstar: -inf < -9.407 < inf | logz: -18.668 +/- 0.144 | dlogz: 3.814 > 0.409]
- 3426it [00:05, 818.66it/s, bound: 6 | nc: 4 | ncall: 17683 | eff(%): 19.375 | loglstar: -inf < -8.757 < inf | logz: -18.023 +/- 0.144 | dlogz: 2.923 > 0.409]
</pre>
- 3426it [00:05, 818.66it/s, bound: 6 | nc: 4 | ncall: 17683 | eff(%): 19.375 | loglstar: -inf < -8.757 < inf | logz: -18.023 +/- 0.144 | dlogz: 2.923 > 0.409]
end{sphinxVerbatim}
3426it [00:05, 818.66it/s, bound: 6 | nc: 4 | ncall: 17683 | eff(%): 19.375 | loglstar: -inf < -8.757 < inf | logz: -18.023 +/- 0.144 | dlogz: 2.923 > 0.409]
- 3529it [00:06, 683.43it/s, bound: 7 | nc: 1 | ncall: 17845 | eff(%): 19.776 | loglstar: -inf < -8.334 < inf | logz: -17.576 +/- 0.143 | dlogz: 2.271 > 0.409]
</pre>
- 3529it [00:06, 683.43it/s, bound: 7 | nc: 1 | ncall: 17845 | eff(%): 19.776 | loglstar: -inf < -8.334 < inf | logz: -17.576 +/- 0.143 | dlogz: 2.271 > 0.409]
end{sphinxVerbatim}
3529it [00:06, 683.43it/s, bound: 7 | nc: 1 | ncall: 17845 | eff(%): 19.776 | loglstar: -inf < -8.334 < inf | logz: -17.576 +/- 0.143 | dlogz: 2.271 > 0.409]
- 3658it [00:06, 805.88it/s, bound: 7 | nc: 6 | ncall: 18034 | eff(%): 20.284 | loglstar: -inf < -7.845 < inf | logz: -17.143 +/- 0.144 | dlogz: 1.626 > 0.409]
</pre>
- 3658it [00:06, 805.88it/s, bound: 7 | nc: 6 | ncall: 18034 | eff(%): 20.284 | loglstar: -inf < -7.845 < inf | logz: -17.143 +/- 0.144 | dlogz: 1.626 > 0.409]
end{sphinxVerbatim}
3658it [00:06, 805.88it/s, bound: 7 | nc: 6 | ncall: 18034 | eff(%): 20.284 | loglstar: -inf < -7.845 < inf | logz: -17.143 +/- 0.144 | dlogz: 1.626 > 0.409]
- 3770it [00:06, 876.19it/s, bound: 7 | nc: 2 | ncall: 18259 | eff(%): 20.647 | loglstar: -inf < -7.533 < inf | logz: -16.853 +/- 0.144 | dlogz: 1.196 > 0.409]
</pre>
- 3770it [00:06, 876.19it/s, bound: 7 | nc: 2 | ncall: 18259 | eff(%): 20.647 | loglstar: -inf < -7.533 < inf | logz: -16.853 +/- 0.144 | dlogz: 1.196 > 0.409]
end{sphinxVerbatim}
3770it [00:06, 876.19it/s, bound: 7 | nc: 2 | ncall: 18259 | eff(%): 20.647 | loglstar: -inf < -7.533 < inf | logz: -16.853 +/- 0.144 | dlogz: 1.196 > 0.409]
- 3873it [00:06, 664.96it/s, bound: 8 | nc: 1 | ncall: 18441 | eff(%): 21.002 | loglstar: -inf < -7.312 < inf | logz: -16.645 +/- 0.144 | dlogz: 0.896 > 0.409]
</pre>
- 3873it [00:06, 664.96it/s, bound: 8 | nc: 1 | ncall: 18441 | eff(%): 21.002 | loglstar: -inf < -7.312 < inf | logz: -16.645 +/- 0.144 | dlogz: 0.896 > 0.409]
end{sphinxVerbatim}
3873it [00:06, 664.96it/s, bound: 8 | nc: 1 | ncall: 18441 | eff(%): 21.002 | loglstar: -inf < -7.312 < inf | logz: -16.645 +/- 0.144 | dlogz: 0.896 > 0.409]
- 4048it [00:06, 882.23it/s, bound: 8 | nc: 1 | ncall: 18720 | eff(%): 21.624 | loglstar: -inf < -7.092 < inf | logz: -16.395 +/- 0.144 | dlogz: 0.547 > 0.409]
</pre>
- 4048it [00:06, 882.23it/s, bound: 8 | nc: 1 | ncall: 18720 | eff(%): 21.624 | loglstar: -inf < -7.092 < inf | logz: -16.395 +/- 0.144 | dlogz: 0.547 > 0.409]
end{sphinxVerbatim}
4048it [00:06, 882.23it/s, bound: 8 | nc: 1 | ncall: 18720 | eff(%): 21.624 | loglstar: -inf < -7.092 < inf | logz: -16.395 +/- 0.144 | dlogz: 0.547 > 0.409]
- 4153it [00:06, 611.51it/s, +400 | bound: 8 | nc: 1 | ncall: 19322 | eff(%): 24.062 | loglstar: -inf < -6.608 < inf | logz: -15.942 +/- 0.144 | dlogz: 0.001 > 0.409]
</pre>
- 4153it [00:06, 611.51it/s, +400 | bound: 8 | nc: 1 | ncall: 19322 | eff(%): 24.062 | loglstar: -inf < -6.608 < inf | logz: -15.942 +/- 0.144 | dlogz: 0.001 > 0.409]
end{sphinxVerbatim}
4153it [00:06, 611.51it/s, +400 | bound: 8 | nc: 1 | ncall: 19322 | eff(%): 24.062 | loglstar: -inf < -6.608 < inf | logz: -15.942 +/- 0.144 | dlogz: 0.001 > 0.409]
22:18:59 INFO fit restored to maximum of posterior sampler_base.py:178
INFO fit restored to maximum of posterior sampler_base.py:178
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | (9.91 +/- 0.16) x 10^-1 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (9.99 +/- 0.05) x 10^-2 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -6.598725 |
| total | -6.598725 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 17.903332 |
| BIC | 19.188914 |
| DIC | 17.335774 |
| PDIC | 2.068719 |
| log(Z) | -6.923581 |
[7]:
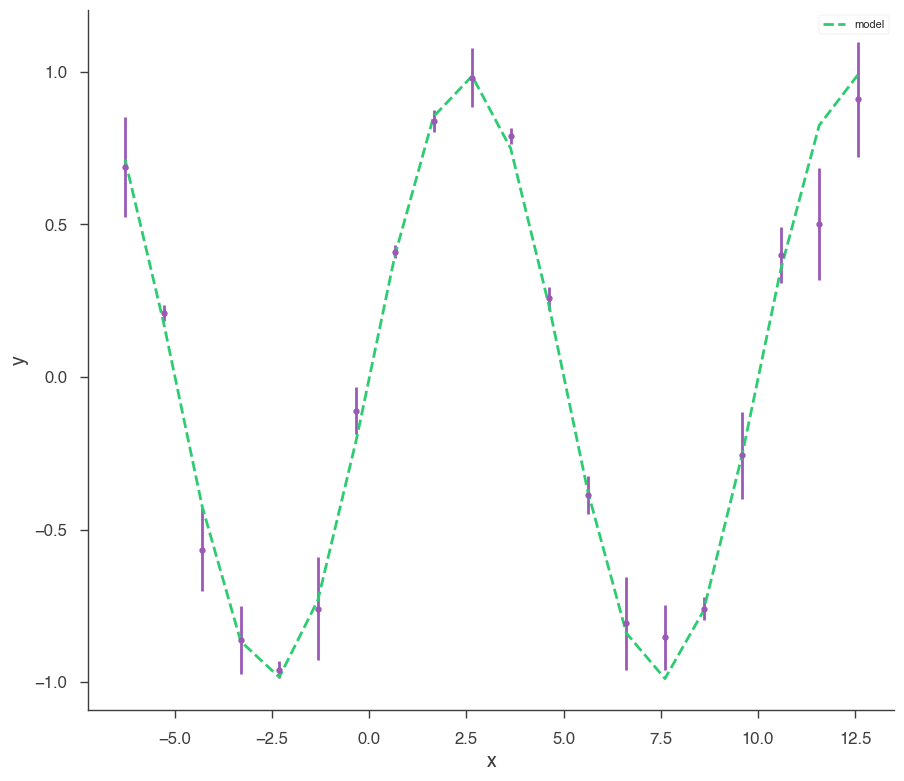
[8]:
bayes_analysis.set_sampler("dynesty_dynamic")
bayes_analysis.sampler.setup(
stop_function=dynesty.utils.old_stopping_function, n_effective=None
)
bayes_analysis.sample()
xyl.plot()
bayes_analysis.results.corner_plot()
22:19:00 INFO sampler set to dynesty_dynamic bayesian_analysis.py:202
- 0it [00:00, ?it/s]
</pre>
- 0it [00:00, ?it/s]
end{sphinxVerbatim}
0it [00:00, ?it/s]
- 1it [00:00, 7.81it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 1 | eff(%): 0.200 | loglstar: -inf < -9730.319 < inf | logz: -9737.229 +/- 0.117 | dlogz: inf > 0.010]
</pre>
- 1it [00:00, 7.81it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 1 | eff(%): 0.200 | loglstar: -inf < -9730.319 < inf | logz: -9737.229 +/- 0.117 | dlogz: inf > 0.010]
end{sphinxVerbatim}
1it [00:00, 7.81it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 1 | eff(%): 0.200 | loglstar: -inf < -9730.319 < inf | logz: -9737.229 +/- 0.117 | dlogz: inf > 0.010]
- 113it [00:00, 593.49it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 122 | eff(%): 18.167 | loglstar: -inf < -5080.547 < inf | logz: -5087.674 +/- 0.119 | dlogz: 5070.630 > 0.010]
</pre>
- 113it [00:00, 593.49it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 122 | eff(%): 18.167 | loglstar: -inf < -5080.547 < inf | logz: -5087.674 +/- 0.119 | dlogz: 5070.630 > 0.010]
end{sphinxVerbatim}
113it [00:00, 593.49it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 122 | eff(%): 18.167 | loglstar: -inf < -5080.547 < inf | logz: -5087.674 +/- 0.119 | dlogz: 5070.630 > 0.010]
- 242it [00:00, 892.34it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 306 | eff(%): 30.025 | loglstar: -inf < -3801.351 < inf | logz: -3808.742 +/- 0.121 | dlogz: 3808.676 > 0.010]
</pre>
- 242it [00:00, 892.34it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 306 | eff(%): 30.025 | loglstar: -inf < -3801.351 < inf | logz: -3808.742 +/- 0.121 | dlogz: 3808.676 > 0.010]
end{sphinxVerbatim}
242it [00:00, 892.34it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 306 | eff(%): 30.025 | loglstar: -inf < -3801.351 < inf | logz: -3808.742 +/- 0.121 | dlogz: 3808.676 > 0.010]
- 337it [00:00, 865.98it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 460 | eff(%): 35.104 | loglstar: -inf < -3336.960 < inf | logz: -3343.678 +/- 0.110 | dlogz: 3335.811 > 0.010]
</pre>
- 337it [00:00, 865.98it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 460 | eff(%): 35.104 | loglstar: -inf < -3336.960 < inf | logz: -3343.678 +/- 0.110 | dlogz: 3335.811 > 0.010]
end{sphinxVerbatim}
337it [00:00, 865.98it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 460 | eff(%): 35.104 | loglstar: -inf < -3336.960 < inf | logz: -3343.678 +/- 0.110 | dlogz: 3335.811 > 0.010]
- 473it [00:00, 1032.56it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 751 | eff(%): 37.810 | loglstar: -inf < -2915.463 < inf | logz: -2921.918 +/- 0.110 | dlogz: 2913.418 > 0.010]
</pre>
- 473it [00:00, 1032.56it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 751 | eff(%): 37.810 | loglstar: -inf < -2915.463 < inf | logz: -2921.918 +/- 0.110 | dlogz: 2913.418 > 0.010]
end{sphinxVerbatim}
473it [00:00, 1032.56it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 751 | eff(%): 37.810 | loglstar: -inf < -2915.463 < inf | logz: -2921.918 +/- 0.110 | dlogz: 2913.418 > 0.010]
- 580it [00:00, 905.09it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 1022 | eff(%): 38.108 | loglstar: -inf < -2593.152 < inf | logz: -2600.827 +/- 0.118 | dlogz: 2594.527 > 0.010]
</pre>
- 580it [00:00, 905.09it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 1022 | eff(%): 38.108 | loglstar: -inf < -2593.152 < inf | logz: -2600.827 +/- 0.118 | dlogz: 2594.527 > 0.010]
end{sphinxVerbatim}
580it [00:00, 905.09it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 1022 | eff(%): 38.108 | loglstar: -inf < -2593.152 < inf | logz: -2600.827 +/- 0.118 | dlogz: 2594.527 > 0.010]
- 685it [00:00, 945.53it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 1397 | eff(%): 36.110 | loglstar: -inf < -2337.326 < inf | logz: -2345.590 +/- 0.128 | dlogz: 2342.345 > 0.010]
</pre>
- 685it [00:00, 945.53it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 1397 | eff(%): 36.110 | loglstar: -inf < -2337.326 < inf | logz: -2345.590 +/- 0.128 | dlogz: 2342.345 > 0.010]
end{sphinxVerbatim}
685it [00:00, 945.53it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 1397 | eff(%): 36.110 | loglstar: -inf < -2337.326 < inf | logz: -2345.590 +/- 0.128 | dlogz: 2342.345 > 0.010]
- 784it [00:01, 655.59it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 1860 | eff(%): 33.220 | loglstar: -inf < -2153.064 < inf | logz: -2161.455 +/- 0.127 | dlogz: 2156.069 > 0.010]
</pre>
- 784it [00:01, 655.59it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 1860 | eff(%): 33.220 | loglstar: -inf < -2153.064 < inf | logz: -2161.455 +/- 0.127 | dlogz: 2156.069 > 0.010]
end{sphinxVerbatim}
784it [00:01, 655.59it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 1860 | eff(%): 33.220 | loglstar: -inf < -2153.064 < inf | logz: -2161.455 +/- 0.127 | dlogz: 2156.069 > 0.010]
- 864it [00:01, 680.34it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 2254 | eff(%): 31.373 | loglstar: -inf < -2008.775 < inf | logz: -2016.491 +/- 0.118 | dlogz: 2008.928 > 0.010]
</pre>
- 864it [00:01, 680.34it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 2254 | eff(%): 31.373 | loglstar: -inf < -2008.775 < inf | logz: -2016.491 +/- 0.118 | dlogz: 2008.928 > 0.010]
end{sphinxVerbatim}
864it [00:01, 680.34it/s, batch: 0 | bound: 0 | nc: 1 | ncall: 2254 | eff(%): 31.373 | loglstar: -inf < -2008.775 < inf | logz: -2016.491 +/- 0.118 | dlogz: 2008.928 > 0.010]
- 943it [00:01, 690.18it/s, batch: 0 | bound: 0 | nc: 9 | ncall: 2648 | eff(%): 29.956 | loglstar: -inf < -1931.436 < inf | logz: -1938.188 +/- 0.110 | dlogz: 1929.962 > 0.010]
</pre>
- 943it [00:01, 690.18it/s, batch: 0 | bound: 0 | nc: 9 | ncall: 2648 | eff(%): 29.956 | loglstar: -inf < -1931.436 < inf | logz: -1938.188 +/- 0.110 | dlogz: 1929.962 > 0.010]
end{sphinxVerbatim}
943it [00:01, 690.18it/s, batch: 0 | bound: 0 | nc: 9 | ncall: 2648 | eff(%): 29.956 | loglstar: -inf < -1931.436 < inf | logz: -1938.188 +/- 0.110 | dlogz: 1929.962 > 0.010]
- 1020it [00:01, 652.97it/s, batch: 0 | bound: 0 | nc: 5 | ncall: 3127 | eff(%): 28.122 | loglstar: -inf < -1610.162 < inf | logz: -1619.108 +/- 0.134 | dlogz: 1626.459 > 0.010]
</pre>
- 1020it [00:01, 652.97it/s, batch: 0 | bound: 0 | nc: 5 | ncall: 3127 | eff(%): 28.122 | loglstar: -inf < -1610.162 < inf | logz: -1619.108 +/- 0.134 | dlogz: 1626.459 > 0.010]
end{sphinxVerbatim}
1020it [00:01, 652.97it/s, batch: 0 | bound: 0 | nc: 5 | ncall: 3127 | eff(%): 28.122 | loglstar: -inf < -1610.162 < inf | logz: -1619.108 +/- 0.134 | dlogz: 1626.459 > 0.010]
- 1091it [00:01, 636.18it/s, batch: 0 | bound: 0 | nc: 3 | ncall: 3599 | eff(%): 26.616 | loglstar: -inf < -1351.994 < inf | logz: -1360.769 +/- 0.126 | dlogz: 1353.604 > 0.010]
</pre>
- 1091it [00:01, 636.18it/s, batch: 0 | bound: 0 | nc: 3 | ncall: 3599 | eff(%): 26.616 | loglstar: -inf < -1351.994 < inf | logz: -1360.769 +/- 0.126 | dlogz: 1353.604 > 0.010]
end{sphinxVerbatim}
1091it [00:01, 636.18it/s, batch: 0 | bound: 0 | nc: 3 | ncall: 3599 | eff(%): 26.616 | loglstar: -inf < -1351.994 < inf | logz: -1360.769 +/- 0.126 | dlogz: 1353.604 > 0.010]
- 1159it [00:01, 579.75it/s, batch: 0 | bound: 0 | nc: 12 | ncall: 4195 | eff(%): 24.686 | loglstar: -inf < -1096.884 < inf | logz: -1106.107 +/- 0.136 | dlogz: 1105.564 > 0.010]
</pre>
- 1159it [00:01, 579.75it/s, batch: 0 | bound: 0 | nc: 12 | ncall: 4195 | eff(%): 24.686 | loglstar: -inf < -1096.884 < inf | logz: -1106.107 +/- 0.136 | dlogz: 1105.564 > 0.010]
end{sphinxVerbatim}
1159it [00:01, 579.75it/s, batch: 0 | bound: 0 | nc: 12 | ncall: 4195 | eff(%): 24.686 | loglstar: -inf < -1096.884 < inf | logz: -1106.107 +/- 0.136 | dlogz: 1105.564 > 0.010]
- 1220it [00:01, 541.08it/s, batch: 0 | bound: 0 | nc: 9 | ncall: 4899 | eff(%): 22.597 | loglstar: -inf < -907.924 < inf | logz: -916.564 +/- 0.127 | dlogz: 908.695 > 0.010]
</pre>
- 1220it [00:01, 541.08it/s, batch: 0 | bound: 0 | nc: 9 | ncall: 4899 | eff(%): 22.597 | loglstar: -inf < -907.924 < inf | logz: -916.564 +/- 0.127 | dlogz: 908.695 > 0.010]
end{sphinxVerbatim}
1220it [00:01, 541.08it/s, batch: 0 | bound: 0 | nc: 9 | ncall: 4899 | eff(%): 22.597 | loglstar: -inf < -907.924 < inf | logz: -916.564 +/- 0.127 | dlogz: 908.695 > 0.010]
- 1277it [00:01, 469.90it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 5640 | eff(%): 20.798 | loglstar: -inf < -788.123 < inf | logz: -796.019 +/- 0.120 | dlogz: 787.342 > 0.010]
</pre>
- 1277it [00:01, 469.90it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 5640 | eff(%): 20.798 | loglstar: -inf < -788.123 < inf | logz: -796.019 +/- 0.120 | dlogz: 787.342 > 0.010]
end{sphinxVerbatim}
1277it [00:01, 469.90it/s, batch: 0 | bound: 0 | nc: 2 | ncall: 5640 | eff(%): 20.798 | loglstar: -inf < -788.123 < inf | logz: -796.019 +/- 0.120 | dlogz: 787.342 > 0.010]
- 1327it [00:02, 449.02it/s, batch: 0 | bound: 0 | nc: 13 | ncall: 6158 | eff(%): 19.931 | loglstar: -inf < -686.807 < inf | logz: -695.597 +/- 0.127 | dlogz: 687.256 > 0.010]
</pre>
- 1327it [00:02, 449.02it/s, batch: 0 | bound: 0 | nc: 13 | ncall: 6158 | eff(%): 19.931 | loglstar: -inf < -686.807 < inf | logz: -695.597 +/- 0.127 | dlogz: 687.256 > 0.010]
end{sphinxVerbatim}
1327it [00:02, 449.02it/s, batch: 0 | bound: 0 | nc: 13 | ncall: 6158 | eff(%): 19.931 | loglstar: -inf < -686.807 < inf | logz: -695.597 +/- 0.127 | dlogz: 687.256 > 0.010]
- 1374it [00:02, 438.54it/s, batch: 0 | bound: 0 | nc: 24 | ncall: 6791 | eff(%): 18.845 | loglstar: -inf < -612.067 < inf | logz: -621.603 +/- 0.135 | dlogz: 614.683 > 0.010]
</pre>
- 1374it [00:02, 438.54it/s, batch: 0 | bound: 0 | nc: 24 | ncall: 6791 | eff(%): 18.845 | loglstar: -inf < -612.067 < inf | logz: -621.603 +/- 0.135 | dlogz: 614.683 > 0.010]
end{sphinxVerbatim}
1374it [00:02, 438.54it/s, batch: 0 | bound: 0 | nc: 24 | ncall: 6791 | eff(%): 18.845 | loglstar: -inf < -612.067 < inf | logz: -621.603 +/- 0.135 | dlogz: 614.683 > 0.010]
- 1419it [00:02, 432.53it/s, batch: 0 | bound: 0 | nc: 17 | ncall: 7445 | eff(%): 17.860 | loglstar: -inf < -542.056 < inf | logz: -550.805 +/- 0.124 | dlogz: 542.076 > 0.010]
</pre>
- 1419it [00:02, 432.53it/s, batch: 0 | bound: 0 | nc: 17 | ncall: 7445 | eff(%): 17.860 | loglstar: -inf < -542.056 < inf | logz: -550.805 +/- 0.124 | dlogz: 542.076 > 0.010]
end{sphinxVerbatim}
1419it [00:02, 432.53it/s, batch: 0 | bound: 0 | nc: 17 | ncall: 7445 | eff(%): 17.860 | loglstar: -inf < -542.056 < inf | logz: -550.805 +/- 0.124 | dlogz: 542.076 > 0.010]
- 1463it [00:02, 353.00it/s, batch: 0 | bound: 0 | nc: 36 | ncall: 8325 | eff(%): 16.578 | loglstar: -inf < -493.076 < inf | logz: -502.111 +/- 0.127 | dlogz: 493.494 > 0.010]
</pre>
- 1463it [00:02, 353.00it/s, batch: 0 | bound: 0 | nc: 36 | ncall: 8325 | eff(%): 16.578 | loglstar: -inf < -493.076 < inf | logz: -502.111 +/- 0.127 | dlogz: 493.494 > 0.010]
end{sphinxVerbatim}
1463it [00:02, 353.00it/s, batch: 0 | bound: 0 | nc: 36 | ncall: 8325 | eff(%): 16.578 | loglstar: -inf < -493.076 < inf | logz: -502.111 +/- 0.127 | dlogz: 493.494 > 0.010]
- 1501it [00:02, 268.52it/s, batch: 0 | bound: 0 | nc: 12 | ncall: 9091 | eff(%): 15.650 | loglstar: -inf < -456.489 < inf | logz: -465.994 +/- 0.131 | dlogz: 457.742 > 0.010]
</pre>
- 1501it [00:02, 268.52it/s, batch: 0 | bound: 0 | nc: 12 | ncall: 9091 | eff(%): 15.650 | loglstar: -inf < -456.489 < inf | logz: -465.994 +/- 0.131 | dlogz: 457.742 > 0.010]
end{sphinxVerbatim}
1501it [00:02, 268.52it/s, batch: 0 | bound: 0 | nc: 12 | ncall: 9091 | eff(%): 15.650 | loglstar: -inf < -456.489 < inf | logz: -465.994 +/- 0.131 | dlogz: 457.742 > 0.010]
- 1532it [00:02, 246.44it/s, batch: 0 | bound: 0 | nc: 76 | ncall: 9848 | eff(%): 14.805 | loglstar: -inf < -434.093 < inf | logz: -442.667 +/- 0.124 | dlogz: 433.465 > 0.010]
</pre>
- 1532it [00:02, 246.44it/s, batch: 0 | bound: 0 | nc: 76 | ncall: 9848 | eff(%): 14.805 | loglstar: -inf < -434.093 < inf | logz: -442.667 +/- 0.124 | dlogz: 433.465 > 0.010]
end{sphinxVerbatim}
1532it [00:02, 246.44it/s, batch: 0 | bound: 0 | nc: 76 | ncall: 9848 | eff(%): 14.805 | loglstar: -inf < -434.093 < inf | logz: -442.667 +/- 0.124 | dlogz: 433.465 > 0.010]
- 1560it [00:03, 237.30it/s, batch: 0 | bound: 0 | nc: 37 | ncall: 10488 | eff(%): 14.197 | loglstar: -inf < -413.986 < inf | logz: -422.557 +/- 0.125 | dlogz: 413.370 > 0.010]
</pre>
- 1560it [00:03, 237.30it/s, batch: 0 | bound: 0 | nc: 37 | ncall: 10488 | eff(%): 14.197 | loglstar: -inf < -413.986 < inf | logz: -422.557 +/- 0.125 | dlogz: 413.370 > 0.010]
end{sphinxVerbatim}
1560it [00:03, 237.30it/s, batch: 0 | bound: 0 | nc: 37 | ncall: 10488 | eff(%): 14.197 | loglstar: -inf < -413.986 < inf | logz: -422.557 +/- 0.125 | dlogz: 413.370 > 0.010]
- 1586it [00:03, 207.64it/s, batch: 0 | bound: 0 | nc: 12 | ncall: 11141 | eff(%): 13.624 | loglstar: -inf < -398.893 < inf | logz: -406.994 +/- 0.122 | dlogz: 397.519 > 0.010]
</pre>
- 1586it [00:03, 207.64it/s, batch: 0 | bound: 0 | nc: 12 | ncall: 11141 | eff(%): 13.624 | loglstar: -inf < -398.893 < inf | logz: -406.994 +/- 0.122 | dlogz: 397.519 > 0.010]
end{sphinxVerbatim}
1586it [00:03, 207.64it/s, batch: 0 | bound: 0 | nc: 12 | ncall: 11141 | eff(%): 13.624 | loglstar: -inf < -398.893 < inf | logz: -406.994 +/- 0.122 | dlogz: 397.519 > 0.010]
- 1609it [00:03, 187.84it/s, batch: 0 | bound: 0 | nc: 37 | ncall: 11940 | eff(%): 12.934 | loglstar: -inf < -383.916 < inf | logz: -393.003 +/- 0.129 | dlogz: 383.864 > 0.010]
</pre>
- 1609it [00:03, 187.84it/s, batch: 0 | bound: 0 | nc: 37 | ncall: 11940 | eff(%): 12.934 | loglstar: -inf < -383.916 < inf | logz: -393.003 +/- 0.129 | dlogz: 383.864 > 0.010]
end{sphinxVerbatim}
1609it [00:03, 187.84it/s, batch: 0 | bound: 0 | nc: 37 | ncall: 11940 | eff(%): 12.934 | loglstar: -inf < -383.916 < inf | logz: -393.003 +/- 0.129 | dlogz: 383.864 > 0.010]
- 1629it [00:03, 189.32it/s, batch: 0 | bound: 0 | nc: 49 | ncall: 12404 | eff(%): 12.624 | loglstar: -inf < -368.271 < inf | logz: -377.174 +/- 0.128 | dlogz: 367.926 > 0.010]
</pre>
- 1629it [00:03, 189.32it/s, batch: 0 | bound: 0 | nc: 49 | ncall: 12404 | eff(%): 12.624 | loglstar: -inf < -368.271 < inf | logz: -377.174 +/- 0.128 | dlogz: 367.926 > 0.010]
end{sphinxVerbatim}
1629it [00:03, 189.32it/s, batch: 0 | bound: 0 | nc: 49 | ncall: 12404 | eff(%): 12.624 | loglstar: -inf < -368.271 < inf | logz: -377.174 +/- 0.128 | dlogz: 367.926 > 0.010]
- 1649it [00:03, 188.16it/s, batch: 0 | bound: 0 | nc: 24 | ncall: 12948 | eff(%): 12.262 | loglstar: -inf < -354.622 < inf | logz: -363.851 +/- 0.126 | dlogz: 354.589 > 0.010]
</pre>
- 1649it [00:03, 188.16it/s, batch: 0 | bound: 0 | nc: 24 | ncall: 12948 | eff(%): 12.262 | loglstar: -inf < -354.622 < inf | logz: -363.851 +/- 0.126 | dlogz: 354.589 > 0.010]
end{sphinxVerbatim}
1649it [00:03, 188.16it/s, batch: 0 | bound: 0 | nc: 24 | ncall: 12948 | eff(%): 12.262 | loglstar: -inf < -354.622 < inf | logz: -363.851 +/- 0.126 | dlogz: 354.589 > 0.010]
- 1669it [00:03, 150.80it/s, batch: 0 | bound: 0 | nc: 52 | ncall: 13406 | eff(%): 12.002 | loglstar: -inf < -342.563 < inf | logz: -351.078 +/- 0.126 | dlogz: 341.491 > 0.010]
</pre>
- 1669it [00:03, 150.80it/s, batch: 0 | bound: 0 | nc: 52 | ncall: 13406 | eff(%): 12.002 | loglstar: -inf < -342.563 < inf | logz: -351.078 +/- 0.126 | dlogz: 341.491 > 0.010]
end{sphinxVerbatim}
1669it [00:03, 150.80it/s, batch: 0 | bound: 0 | nc: 52 | ncall: 13406 | eff(%): 12.002 | loglstar: -inf < -342.563 < inf | logz: -351.078 +/- 0.126 | dlogz: 341.491 > 0.010]
- 1695it [00:03, 172.02it/s, batch: 0 | bound: 0 | nc: 57 | ncall: 14082 | eff(%): 11.624 | loglstar: -inf < -328.462 < inf | logz: -337.273 +/- 0.125 | dlogz: 327.724 > 0.010]
</pre>
- 1695it [00:03, 172.02it/s, batch: 0 | bound: 0 | nc: 57 | ncall: 14082 | eff(%): 11.624 | loglstar: -inf < -328.462 < inf | logz: -337.273 +/- 0.125 | dlogz: 327.724 > 0.010]
end{sphinxVerbatim}
1695it [00:03, 172.02it/s, batch: 0 | bound: 0 | nc: 57 | ncall: 14082 | eff(%): 11.624 | loglstar: -inf < -328.462 < inf | logz: -337.273 +/- 0.125 | dlogz: 327.724 > 0.010]
- 1718it [00:04, 184.08it/s, batch: 0 | bound: 0 | nc: 13 | ncall: 14641 | eff(%): 11.347 | loglstar: -inf < -316.280 < inf | logz: -325.024 +/- 0.127 | dlogz: 315.394 > 0.010]
</pre>
- 1718it [00:04, 184.08it/s, batch: 0 | bound: 0 | nc: 13 | ncall: 14641 | eff(%): 11.347 | loglstar: -inf < -316.280 < inf | logz: -325.024 +/- 0.127 | dlogz: 315.394 > 0.010]
end{sphinxVerbatim}
1718it [00:04, 184.08it/s, batch: 0 | bound: 0 | nc: 13 | ncall: 14641 | eff(%): 11.347 | loglstar: -inf < -316.280 < inf | logz: -325.024 +/- 0.127 | dlogz: 315.394 > 0.010]
- 1738it [00:04, 153.34it/s, batch: 0 | bound: 0 | nc: 134 | ncall: 15722 | eff(%): 10.714 | loglstar: -inf < -308.130 < inf | logz: -317.159 +/- 0.128 | dlogz: 307.722 > 0.010]
</pre>
- 1738it [00:04, 153.34it/s, batch: 0 | bound: 0 | nc: 134 | ncall: 15722 | eff(%): 10.714 | loglstar: -inf < -308.130 < inf | logz: -317.159 +/- 0.128 | dlogz: 307.722 > 0.010]
end{sphinxVerbatim}
1738it [00:04, 153.34it/s, batch: 0 | bound: 0 | nc: 134 | ncall: 15722 | eff(%): 10.714 | loglstar: -inf < -308.130 < inf | logz: -317.159 +/- 0.128 | dlogz: 307.722 > 0.010]
- 1756it [00:04, 145.97it/s, batch: 0 | bound: 0 | nc: 31 | ncall: 16474 | eff(%): 10.345 | loglstar: -inf < -300.709 < inf | logz: -309.566 +/- 0.125 | dlogz: 299.845 > 0.010]
</pre>
- 1756it [00:04, 145.97it/s, batch: 0 | bound: 0 | nc: 31 | ncall: 16474 | eff(%): 10.345 | loglstar: -inf < -300.709 < inf | logz: -309.566 +/- 0.125 | dlogz: 299.845 > 0.010]
end{sphinxVerbatim}
1756it [00:04, 145.97it/s, batch: 0 | bound: 0 | nc: 31 | ncall: 16474 | eff(%): 10.345 | loglstar: -inf < -300.709 < inf | logz: -309.566 +/- 0.125 | dlogz: 299.845 > 0.010]
- 1772it [00:04, 126.42it/s, batch: 0 | bound: 0 | nc: 13 | ncall: 17213 | eff(%): 10.004 | loglstar: -inf < -292.034 < inf | logz: -301.281 +/- 0.127 | dlogz: 291.687 > 0.010]
</pre>
- 1772it [00:04, 126.42it/s, batch: 0 | bound: 0 | nc: 13 | ncall: 17213 | eff(%): 10.004 | loglstar: -inf < -292.034 < inf | logz: -301.281 +/- 0.127 | dlogz: 291.687 > 0.010]
end{sphinxVerbatim}
1772it [00:04, 126.42it/s, batch: 0 | bound: 0 | nc: 13 | ncall: 17213 | eff(%): 10.004 | loglstar: -inf < -292.034 < inf | logz: -301.281 +/- 0.127 | dlogz: 291.687 > 0.010]
- 1786it [00:04, 104.43it/s, batch: 0 | bound: 1 | nc: 1 | ncall: 17263 | eff(%): 10.055 | loglstar: -inf < -282.299 < inf | logz: -291.715 +/- 0.133 | dlogz: 282.441 > 0.010]
</pre>
- 1786it [00:04, 104.43it/s, batch: 0 | bound: 1 | nc: 1 | ncall: 17263 | eff(%): 10.055 | loglstar: -inf < -282.299 < inf | logz: -291.715 +/- 0.133 | dlogz: 282.441 > 0.010]
end{sphinxVerbatim}
1786it [00:04, 104.43it/s, batch: 0 | bound: 1 | nc: 1 | ncall: 17263 | eff(%): 10.055 | loglstar: -inf < -282.299 < inf | logz: -291.715 +/- 0.133 | dlogz: 282.441 > 0.010]
- 1945it [00:04, 399.57it/s, batch: 0 | bound: 1 | nc: 1 | ncall: 17479 | eff(%): 10.818 | loglstar: -inf < -215.012 < inf | logz: -224.246 +/- 0.128 | dlogz: 214.189 > 0.010]
</pre>
- 1945it [00:04, 399.57it/s, batch: 0 | bound: 1 | nc: 1 | ncall: 17479 | eff(%): 10.818 | loglstar: -inf < -215.012 < inf | logz: -224.246 +/- 0.128 | dlogz: 214.189 > 0.010]
end{sphinxVerbatim}
1945it [00:04, 399.57it/s, batch: 0 | bound: 1 | nc: 1 | ncall: 17479 | eff(%): 10.818 | loglstar: -inf < -215.012 < inf | logz: -224.246 +/- 0.128 | dlogz: 214.189 > 0.010]
- 2052it [00:04, 546.60it/s, batch: 0 | bound: 1 | nc: 3 | ncall: 17652 | eff(%): 11.305 | loglstar: -inf < -172.242 < inf | logz: -181.162 +/- 0.129 | dlogz: 170.684 > 0.010]
</pre>
- 2052it [00:04, 546.60it/s, batch: 0 | bound: 1 | nc: 3 | ncall: 17652 | eff(%): 11.305 | loglstar: -inf < -172.242 < inf | logz: -181.162 +/- 0.129 | dlogz: 170.684 > 0.010]
end{sphinxVerbatim}
2052it [00:04, 546.60it/s, batch: 0 | bound: 1 | nc: 3 | ncall: 17652 | eff(%): 11.305 | loglstar: -inf < -172.242 < inf | logz: -181.162 +/- 0.129 | dlogz: 170.684 > 0.010]
- 2183it [00:05, 727.13it/s, batch: 0 | bound: 1 | nc: 8 | ncall: 17958 | eff(%): 11.827 | loglstar: -inf < -137.814 < inf | logz: -147.110 +/- 0.127 | dlogz: 136.396 > 0.010]
</pre>
- 2183it [00:05, 727.13it/s, batch: 0 | bound: 1 | nc: 8 | ncall: 17958 | eff(%): 11.827 | loglstar: -inf < -137.814 < inf | logz: -147.110 +/- 0.127 | dlogz: 136.396 > 0.010]
end{sphinxVerbatim}
2183it [00:05, 727.13it/s, batch: 0 | bound: 1 | nc: 8 | ncall: 17958 | eff(%): 11.827 | loglstar: -inf < -137.814 < inf | logz: -147.110 +/- 0.127 | dlogz: 136.396 > 0.010]
- 2272it [00:05, 565.49it/s, batch: 0 | bound: 2 | nc: 1 | ncall: 18081 | eff(%): 12.228 | loglstar: -inf < -118.898 < inf | logz: -127.978 +/- 0.129 | dlogz: 117.020 > 0.010]
</pre>
- 2272it [00:05, 565.49it/s, batch: 0 | bound: 2 | nc: 1 | ncall: 18081 | eff(%): 12.228 | loglstar: -inf < -118.898 < inf | logz: -127.978 +/- 0.129 | dlogz: 117.020 > 0.010]
end{sphinxVerbatim}
2272it [00:05, 565.49it/s, batch: 0 | bound: 2 | nc: 1 | ncall: 18081 | eff(%): 12.228 | loglstar: -inf < -118.898 < inf | logz: -127.978 +/- 0.129 | dlogz: 117.020 > 0.010]
- 2378it [00:05, 554.25it/s, batch: 0 | bound: 2 | nc: 1 | ncall: 18216 | eff(%): 12.706 | loglstar: -inf < -101.052 < inf | logz: -110.581 +/- 0.128 | dlogz: 99.414 > 0.010]
</pre>
- 2378it [00:05, 554.25it/s, batch: 0 | bound: 2 | nc: 1 | ncall: 18216 | eff(%): 12.706 | loglstar: -inf < -101.052 < inf | logz: -110.581 +/- 0.128 | dlogz: 99.414 > 0.010]
end{sphinxVerbatim}
2378it [00:05, 554.25it/s, batch: 0 | bound: 2 | nc: 1 | ncall: 18216 | eff(%): 12.706 | loglstar: -inf < -101.052 < inf | logz: -110.581 +/- 0.128 | dlogz: 99.414 > 0.010]
- 2505it [00:05, 696.96it/s, batch: 0 | bound: 2 | nc: 5 | ncall: 18430 | eff(%): 13.233 | loglstar: -inf < -77.910 < inf | logz: -87.176 +/- 0.128 | dlogz: 75.695 > 0.010]
</pre>
- 2505it [00:05, 696.96it/s, batch: 0 | bound: 2 | nc: 5 | ncall: 18430 | eff(%): 13.233 | loglstar: -inf < -77.910 < inf | logz: -87.176 +/- 0.128 | dlogz: 75.695 > 0.010]
end{sphinxVerbatim}
2505it [00:05, 696.96it/s, batch: 0 | bound: 2 | nc: 5 | ncall: 18430 | eff(%): 13.233 | loglstar: -inf < -77.910 < inf | logz: -87.176 +/- 0.128 | dlogz: 75.695 > 0.010]
- 2610it [00:05, 774.79it/s, batch: 0 | bound: 2 | nc: 2 | ncall: 18662 | eff(%): 13.621 | loglstar: -inf < -66.910 < inf | logz: -75.557 +/- 0.125 | dlogz: 63.777 > 0.010]
</pre>
- 2610it [00:05, 774.79it/s, batch: 0 | bound: 2 | nc: 2 | ncall: 18662 | eff(%): 13.621 | loglstar: -inf < -66.910 < inf | logz: -75.557 +/- 0.125 | dlogz: 63.777 > 0.010]
end{sphinxVerbatim}
2610it [00:05, 774.79it/s, batch: 0 | bound: 2 | nc: 2 | ncall: 18662 | eff(%): 13.621 | loglstar: -inf < -66.910 < inf | logz: -75.557 +/- 0.125 | dlogz: 63.777 > 0.010]
- 2701it [00:06, 516.58it/s, batch: 0 | bound: 3 | nc: 2 | ncall: 18819 | eff(%): 13.981 | loglstar: -inf < -57.586 < inf | logz: -67.174 +/- 0.127 | dlogz: 55.259 > 0.010]
</pre>
- 2701it [00:06, 516.58it/s, batch: 0 | bound: 3 | nc: 2 | ncall: 18819 | eff(%): 13.981 | loglstar: -inf < -57.586 < inf | logz: -67.174 +/- 0.127 | dlogz: 55.259 > 0.010]
end{sphinxVerbatim}
2701it [00:06, 516.58it/s, batch: 0 | bound: 3 | nc: 2 | ncall: 18819 | eff(%): 13.981 | loglstar: -inf < -57.586 < inf | logz: -67.174 +/- 0.127 | dlogz: 55.259 > 0.010]
- 2844it [00:06, 680.65it/s, batch: 0 | bound: 3 | nc: 1 | ncall: 19015 | eff(%): 14.573 | loglstar: -inf < -43.104 < inf | logz: -52.490 +/- 0.130 | dlogz: 40.265 > 0.010]
</pre>
- 2844it [00:06, 680.65it/s, batch: 0 | bound: 3 | nc: 1 | ncall: 19015 | eff(%): 14.573 | loglstar: -inf < -43.104 < inf | logz: -52.490 +/- 0.130 | dlogz: 40.265 > 0.010]
end{sphinxVerbatim}
2844it [00:06, 680.65it/s, batch: 0 | bound: 3 | nc: 1 | ncall: 19015 | eff(%): 14.573 | loglstar: -inf < -43.104 < inf | logz: -52.490 +/- 0.130 | dlogz: 40.265 > 0.010]
- 2986it [00:06, 830.88it/s, batch: 0 | bound: 3 | nc: 4 | ncall: 19277 | eff(%): 15.098 | loglstar: -inf < -34.120 < inf | logz: -43.453 +/- 0.128 | dlogz: 30.920 > 0.010]
</pre>
- 2986it [00:06, 830.88it/s, batch: 0 | bound: 3 | nc: 4 | ncall: 19277 | eff(%): 15.098 | loglstar: -inf < -34.120 < inf | logz: -43.453 +/- 0.128 | dlogz: 30.920 > 0.010]
end{sphinxVerbatim}
2986it [00:06, 830.88it/s, batch: 0 | bound: 3 | nc: 4 | ncall: 19277 | eff(%): 15.098 | loglstar: -inf < -34.120 < inf | logz: -43.453 +/- 0.128 | dlogz: 30.920 > 0.010]
- 3094it [00:06, 610.01it/s, batch: 0 | bound: 4 | nc: 1 | ncall: 19527 | eff(%): 15.449 | loglstar: -inf < -28.551 < inf | logz: -38.020 +/- 0.130 | dlogz: 25.269 > 0.010]
</pre>
- 3094it [00:06, 610.01it/s, batch: 0 | bound: 4 | nc: 1 | ncall: 19527 | eff(%): 15.449 | loglstar: -inf < -28.551 < inf | logz: -38.020 +/- 0.130 | dlogz: 25.269 > 0.010]
end{sphinxVerbatim}
3094it [00:06, 610.01it/s, batch: 0 | bound: 4 | nc: 1 | ncall: 19527 | eff(%): 15.449 | loglstar: -inf < -28.551 < inf | logz: -38.020 +/- 0.130 | dlogz: 25.269 > 0.010]
- 3218it [00:06, 723.75it/s, batch: 0 | bound: 4 | nc: 1 | ncall: 19675 | eff(%): 15.950 | loglstar: -inf < -24.292 < inf | logz: -33.556 +/- 0.128 | dlogz: 20.550 > 0.010]
</pre>
- 3218it [00:06, 723.75it/s, batch: 0 | bound: 4 | nc: 1 | ncall: 19675 | eff(%): 15.950 | loglstar: -inf < -24.292 < inf | logz: -33.556 +/- 0.128 | dlogz: 20.550 > 0.010]
end{sphinxVerbatim}
3218it [00:06, 723.75it/s, batch: 0 | bound: 4 | nc: 1 | ncall: 19675 | eff(%): 15.950 | loglstar: -inf < -24.292 < inf | logz: -33.556 +/- 0.128 | dlogz: 20.550 > 0.010]
- 3332it [00:06, 807.85it/s, batch: 0 | bound: 4 | nc: 3 | ncall: 19849 | eff(%): 16.374 | loglstar: -inf < -20.661 < inf | logz: -29.950 +/- 0.128 | dlogz: 16.710 > 0.010]
</pre>
- 3332it [00:06, 807.85it/s, batch: 0 | bound: 4 | nc: 3 | ncall: 19849 | eff(%): 16.374 | loglstar: -inf < -20.661 < inf | logz: -29.950 +/- 0.128 | dlogz: 16.710 > 0.010]
end{sphinxVerbatim}
3332it [00:06, 807.85it/s, batch: 0 | bound: 4 | nc: 3 | ncall: 19849 | eff(%): 16.374 | loglstar: -inf < -20.661 < inf | logz: -29.950 +/- 0.128 | dlogz: 16.710 > 0.010]
- 3464it [00:06, 923.55it/s, batch: 0 | bound: 4 | nc: 1 | ncall: 20108 | eff(%): 16.809 | loglstar: -inf < -17.694 < inf | logz: -26.931 +/- 0.128 | dlogz: 13.420 > 0.010]
</pre>
- 3464it [00:06, 923.55it/s, batch: 0 | bound: 4 | nc: 1 | ncall: 20108 | eff(%): 16.809 | loglstar: -inf < -17.694 < inf | logz: -26.931 +/- 0.128 | dlogz: 13.420 > 0.010]
end{sphinxVerbatim}
3464it [00:06, 923.55it/s, batch: 0 | bound: 4 | nc: 1 | ncall: 20108 | eff(%): 16.809 | loglstar: -inf < -17.694 < inf | logz: -26.931 +/- 0.128 | dlogz: 13.420 > 0.010]
- 3575it [00:07, 635.63it/s, batch: 0 | bound: 5 | nc: 1 | ncall: 20305 | eff(%): 17.183 | loglstar: -inf < -15.615 < inf | logz: -24.865 +/- 0.129 | dlogz: 11.129 > 0.010]
</pre>
- 3575it [00:07, 635.63it/s, batch: 0 | bound: 5 | nc: 1 | ncall: 20305 | eff(%): 17.183 | loglstar: -inf < -15.615 < inf | logz: -24.865 +/- 0.129 | dlogz: 11.129 > 0.010]
end{sphinxVerbatim}
3575it [00:07, 635.63it/s, batch: 0 | bound: 5 | nc: 1 | ncall: 20305 | eff(%): 17.183 | loglstar: -inf < -15.615 < inf | logz: -24.865 +/- 0.129 | dlogz: 11.129 > 0.010]
- 3696it [00:07, 742.07it/s, batch: 0 | bound: 5 | nc: 1 | ncall: 20468 | eff(%): 17.627 | loglstar: -inf < -13.534 < inf | logz: -22.945 +/- 0.129 | dlogz: 8.966 > 0.010]
</pre>
- 3696it [00:07, 742.07it/s, batch: 0 | bound: 5 | nc: 1 | ncall: 20468 | eff(%): 17.627 | loglstar: -inf < -13.534 < inf | logz: -22.945 +/- 0.129 | dlogz: 8.966 > 0.010]
end{sphinxVerbatim}
3696it [00:07, 742.07it/s, batch: 0 | bound: 5 | nc: 1 | ncall: 20468 | eff(%): 17.627 | loglstar: -inf < -13.534 < inf | logz: -22.945 +/- 0.129 | dlogz: 8.966 > 0.010]
- 3828it [00:07, 862.92it/s, batch: 0 | bound: 5 | nc: 2 | ncall: 20695 | eff(%): 18.061 | loglstar: -inf < -11.966 < inf | logz: -21.271 +/- 0.129 | dlogz: 7.024 > 0.010]
</pre>
- 3828it [00:07, 862.92it/s, batch: 0 | bound: 5 | nc: 2 | ncall: 20695 | eff(%): 18.061 | loglstar: -inf < -11.966 < inf | logz: -21.271 +/- 0.129 | dlogz: 7.024 > 0.010]
end{sphinxVerbatim}
3828it [00:07, 862.92it/s, batch: 0 | bound: 5 | nc: 2 | ncall: 20695 | eff(%): 18.061 | loglstar: -inf < -11.966 < inf | logz: -21.271 +/- 0.129 | dlogz: 7.024 > 0.010]
- 3963it [00:07, 975.31it/s, batch: 0 | bound: 5 | nc: 4 | ncall: 20981 | eff(%): 18.449 | loglstar: -inf < -10.793 < inf | logz: -20.070 +/- 0.129 | dlogz: 5.554 > 0.010]
</pre>
- 3963it [00:07, 975.31it/s, batch: 0 | bound: 5 | nc: 4 | ncall: 20981 | eff(%): 18.449 | loglstar: -inf < -10.793 < inf | logz: -20.070 +/- 0.129 | dlogz: 5.554 > 0.010]
end{sphinxVerbatim}
3963it [00:07, 975.31it/s, batch: 0 | bound: 5 | nc: 4 | ncall: 20981 | eff(%): 18.449 | loglstar: -inf < -10.793 < inf | logz: -20.070 +/- 0.129 | dlogz: 5.554 > 0.010]
- 4080it [00:07, 733.93it/s, batch: 0 | bound: 6 | nc: 1 | ncall: 21134 | eff(%): 18.859 | loglstar: -inf < -9.762 < inf | logz: -19.212 +/- 0.129 | dlogz: 4.469 > 0.010]
</pre>
- 4080it [00:07, 733.93it/s, batch: 0 | bound: 6 | nc: 1 | ncall: 21134 | eff(%): 18.859 | loglstar: -inf < -9.762 < inf | logz: -19.212 +/- 0.129 | dlogz: 4.469 > 0.010]
end{sphinxVerbatim}
4080it [00:07, 733.93it/s, batch: 0 | bound: 6 | nc: 1 | ncall: 21134 | eff(%): 18.859 | loglstar: -inf < -9.762 < inf | logz: -19.212 +/- 0.129 | dlogz: 4.469 > 0.010]
- 4231it [00:07, 890.85it/s, batch: 0 | bound: 6 | nc: 2 | ncall: 21414 | eff(%): 19.307 | loglstar: -inf < -8.831 < inf | logz: -18.255 +/- 0.130 | dlogz: 3.237 > 0.010]
</pre>
- 4231it [00:07, 890.85it/s, batch: 0 | bound: 6 | nc: 2 | ncall: 21414 | eff(%): 19.307 | loglstar: -inf < -8.831 < inf | logz: -18.255 +/- 0.130 | dlogz: 3.237 > 0.010]
end{sphinxVerbatim}
4231it [00:07, 890.85it/s, batch: 0 | bound: 6 | nc: 2 | ncall: 21414 | eff(%): 19.307 | loglstar: -inf < -8.831 < inf | logz: -18.255 +/- 0.130 | dlogz: 3.237 > 0.010]
- 4343it [00:07, 886.09it/s, batch: 0 | bound: 6 | nc: 2 | ncall: 21669 | eff(%): 19.590 | loglstar: -inf < -8.384 < inf | logz: -17.743 +/- 0.130 | dlogz: 2.542 > 0.010]
</pre>
- 4343it [00:07, 886.09it/s, batch: 0 | bound: 6 | nc: 2 | ncall: 21669 | eff(%): 19.590 | loglstar: -inf < -8.384 < inf | logz: -17.743 +/- 0.130 | dlogz: 2.542 > 0.010]
end{sphinxVerbatim}
4343it [00:07, 886.09it/s, batch: 0 | bound: 6 | nc: 2 | ncall: 21669 | eff(%): 19.590 | loglstar: -inf < -8.384 < inf | logz: -17.743 +/- 0.130 | dlogz: 2.542 > 0.010]
- 4448it [00:08, 511.59it/s, batch: 0 | bound: 7 | nc: 1 | ncall: 21850 | eff(%): 19.902 | loglstar: -inf < -8.020 < inf | logz: -17.376 +/- 0.129 | dlogz: 2.023 > 0.010]
</pre>
- 4448it [00:08, 511.59it/s, batch: 0 | bound: 7 | nc: 1 | ncall: 21850 | eff(%): 19.902 | loglstar: -inf < -8.020 < inf | logz: -17.376 +/- 0.129 | dlogz: 2.023 > 0.010]
end{sphinxVerbatim}
4448it [00:08, 511.59it/s, batch: 0 | bound: 7 | nc: 1 | ncall: 21850 | eff(%): 19.902 | loglstar: -inf < -8.020 < inf | logz: -17.376 +/- 0.129 | dlogz: 2.023 > 0.010]
- 4529it [00:08, 349.32it/s, batch: 0 | bound: 7 | nc: 1 | ncall: 21966 | eff(%): 20.159 | loglstar: -inf < -7.766 < inf | logz: -17.132 +/- 0.129 | dlogz: 1.681 > 0.010]
</pre>
- 4529it [00:08, 349.32it/s, batch: 0 | bound: 7 | nc: 1 | ncall: 21966 | eff(%): 20.159 | loglstar: -inf < -7.766 < inf | logz: -17.132 +/- 0.129 | dlogz: 1.681 > 0.010]
end{sphinxVerbatim}
4529it [00:08, 349.32it/s, batch: 0 | bound: 7 | nc: 1 | ncall: 21966 | eff(%): 20.159 | loglstar: -inf < -7.766 < inf | logz: -17.132 +/- 0.129 | dlogz: 1.681 > 0.010]
- 4672it [00:08, 480.71it/s, batch: 0 | bound: 7 | nc: 2 | ncall: 22220 | eff(%): 20.563 | loglstar: -inf < -7.485 < inf | logz: -16.798 +/- 0.129 | dlogz: 1.211 > 0.010]
</pre>
- 4672it [00:08, 480.71it/s, batch: 0 | bound: 7 | nc: 2 | ncall: 22220 | eff(%): 20.563 | loglstar: -inf < -7.485 < inf | logz: -16.798 +/- 0.129 | dlogz: 1.211 > 0.010]
end{sphinxVerbatim}
4672it [00:08, 480.71it/s, batch: 0 | bound: 7 | nc: 2 | ncall: 22220 | eff(%): 20.563 | loglstar: -inf < -7.485 < inf | logz: -16.798 +/- 0.129 | dlogz: 1.211 > 0.010]
- 4793it [00:09, 433.70it/s, batch: 0 | bound: 8 | nc: 1 | ncall: 22511 | eff(%): 20.829 | loglstar: -inf < -7.294 < inf | logz: -16.590 +/- 0.129 | dlogz: 0.917 > 0.010]
</pre>
- 4793it [00:09, 433.70it/s, batch: 0 | bound: 8 | nc: 1 | ncall: 22511 | eff(%): 20.829 | loglstar: -inf < -7.294 < inf | logz: -16.590 +/- 0.129 | dlogz: 0.917 > 0.010]
end{sphinxVerbatim}
4793it [00:09, 433.70it/s, batch: 0 | bound: 8 | nc: 1 | ncall: 22511 | eff(%): 20.829 | loglstar: -inf < -7.294 < inf | logz: -16.590 +/- 0.129 | dlogz: 0.917 > 0.010]
- 4940it [00:09, 573.21it/s, batch: 0 | bound: 8 | nc: 5 | ncall: 22688 | eff(%): 21.304 | loglstar: -inf < -7.122 < inf | logz: -16.400 +/- 0.129 | dlogz: 0.656 > 0.010]
</pre>
- 4940it [00:09, 573.21it/s, batch: 0 | bound: 8 | nc: 5 | ncall: 22688 | eff(%): 21.304 | loglstar: -inf < -7.122 < inf | logz: -16.400 +/- 0.129 | dlogz: 0.656 > 0.010]
end{sphinxVerbatim}
4940it [00:09, 573.21it/s, batch: 0 | bound: 8 | nc: 5 | ncall: 22688 | eff(%): 21.304 | loglstar: -inf < -7.122 < inf | logz: -16.400 +/- 0.129 | dlogz: 0.656 > 0.010]
- 5057it [00:09, 669.38it/s, batch: 0 | bound: 8 | nc: 2 | ncall: 22887 | eff(%): 21.623 | loglstar: -inf < -7.019 < inf | logz: -16.285 +/- 0.129 | dlogz: 0.503 > 0.010]
</pre>
- 5057it [00:09, 669.38it/s, batch: 0 | bound: 8 | nc: 2 | ncall: 22887 | eff(%): 21.623 | loglstar: -inf < -7.019 < inf | logz: -16.285 +/- 0.129 | dlogz: 0.503 > 0.010]
end{sphinxVerbatim}
5057it [00:09, 669.38it/s, batch: 0 | bound: 8 | nc: 2 | ncall: 22887 | eff(%): 21.623 | loglstar: -inf < -7.019 < inf | logz: -16.285 +/- 0.129 | dlogz: 0.503 > 0.010]
- 5185it [00:09, 780.90it/s, batch: 0 | bound: 8 | nc: 3 | ncall: 23173 | eff(%): 21.903 | loglstar: -inf < -6.926 < inf | logz: -16.188 +/- 0.129 | dlogz: 0.378 > 0.010]
</pre>
- 5185it [00:09, 780.90it/s, batch: 0 | bound: 8 | nc: 3 | ncall: 23173 | eff(%): 21.903 | loglstar: -inf < -6.926 < inf | logz: -16.188 +/- 0.129 | dlogz: 0.378 > 0.010]
end{sphinxVerbatim}
5185it [00:09, 780.90it/s, batch: 0 | bound: 8 | nc: 3 | ncall: 23173 | eff(%): 21.903 | loglstar: -inf < -6.926 < inf | logz: -16.188 +/- 0.129 | dlogz: 0.378 > 0.010]
- 5293it [00:09, 544.92it/s, batch: 0 | bound: 9 | nc: 1 | ncall: 23336 | eff(%): 22.206 | loglstar: -inf < -6.872 < inf | logz: -16.123 +/- 0.129 | dlogz: 0.298 > 0.010]
</pre>
- 5293it [00:09, 544.92it/s, batch: 0 | bound: 9 | nc: 1 | ncall: 23336 | eff(%): 22.206 | loglstar: -inf < -6.872 < inf | logz: -16.123 +/- 0.129 | dlogz: 0.298 > 0.010]
end{sphinxVerbatim}
5293it [00:09, 544.92it/s, batch: 0 | bound: 9 | nc: 1 | ncall: 23336 | eff(%): 22.206 | loglstar: -inf < -6.872 < inf | logz: -16.123 +/- 0.129 | dlogz: 0.298 > 0.010]
- 5431it [00:10, 680.29it/s, batch: 0 | bound: 9 | nc: 1 | ncall: 23514 | eff(%): 22.616 | loglstar: -inf < -6.815 < inf | logz: -16.060 +/- 0.129 | dlogz: 0.221 > 0.010]
</pre>
- 5431it [00:10, 680.29it/s, batch: 0 | bound: 9 | nc: 1 | ncall: 23514 | eff(%): 22.616 | loglstar: -inf < -6.815 < inf | logz: -16.060 +/- 0.129 | dlogz: 0.221 > 0.010]
end{sphinxVerbatim}
5431it [00:10, 680.29it/s, batch: 0 | bound: 9 | nc: 1 | ncall: 23514 | eff(%): 22.616 | loglstar: -inf < -6.815 < inf | logz: -16.060 +/- 0.129 | dlogz: 0.221 > 0.010]
- 5555it [00:10, 784.11it/s, batch: 0 | bound: 9 | nc: 3 | ncall: 23727 | eff(%): 22.929 | loglstar: -inf < -6.772 < inf | logz: -16.016 +/- 0.129 | dlogz: 0.169 > 0.010]
</pre>
- 5555it [00:10, 784.11it/s, batch: 0 | bound: 9 | nc: 3 | ncall: 23727 | eff(%): 22.929 | loglstar: -inf < -6.772 < inf | logz: -16.016 +/- 0.129 | dlogz: 0.169 > 0.010]
end{sphinxVerbatim}
5555it [00:10, 784.11it/s, batch: 0 | bound: 9 | nc: 3 | ncall: 23727 | eff(%): 22.929 | loglstar: -inf < -6.772 < inf | logz: -16.016 +/- 0.129 | dlogz: 0.169 > 0.010]
- 5674it [00:10, 870.05it/s, batch: 0 | bound: 9 | nc: 1 | ncall: 23980 | eff(%): 23.178 | loglstar: -inf < -6.737 < inf | logz: -15.982 +/- 0.129 | dlogz: 0.132 > 0.010]
</pre>
- 5674it [00:10, 870.05it/s, batch: 0 | bound: 9 | nc: 1 | ncall: 23980 | eff(%): 23.178 | loglstar: -inf < -6.737 < inf | logz: -15.982 +/- 0.129 | dlogz: 0.132 > 0.010]
end{sphinxVerbatim}
5674it [00:10, 870.05it/s, batch: 0 | bound: 9 | nc: 1 | ncall: 23980 | eff(%): 23.178 | loglstar: -inf < -6.737 < inf | logz: -15.982 +/- 0.129 | dlogz: 0.132 > 0.010]
- 5785it [00:10, 688.75it/s, batch: 0 | bound: 10 | nc: 1 | ncall: 24134 | eff(%): 23.484 | loglstar: -inf < -6.712 < inf | logz: -15.958 +/- 0.129 | dlogz: 0.104 > 0.010]
</pre>
- 5785it [00:10, 688.75it/s, batch: 0 | bound: 10 | nc: 1 | ncall: 24134 | eff(%): 23.484 | loglstar: -inf < -6.712 < inf | logz: -15.958 +/- 0.129 | dlogz: 0.104 > 0.010]
end{sphinxVerbatim}
5785it [00:10, 688.75it/s, batch: 0 | bound: 10 | nc: 1 | ncall: 24134 | eff(%): 23.484 | loglstar: -inf < -6.712 < inf | logz: -15.958 +/- 0.129 | dlogz: 0.104 > 0.010]
- 5929it [00:10, 837.45it/s, batch: 0 | bound: 10 | nc: 1 | ncall: 24349 | eff(%): 23.860 | loglstar: -inf < -6.687 < inf | logz: -15.933 +/- 0.129 | dlogz: 0.078 > 0.010]
</pre>
- 5929it [00:10, 837.45it/s, batch: 0 | bound: 10 | nc: 1 | ncall: 24349 | eff(%): 23.860 | loglstar: -inf < -6.687 < inf | logz: -15.933 +/- 0.129 | dlogz: 0.078 > 0.010]
end{sphinxVerbatim}
5929it [00:10, 837.45it/s, batch: 0 | bound: 10 | nc: 1 | ncall: 24349 | eff(%): 23.860 | loglstar: -inf < -6.687 < inf | logz: -15.933 +/- 0.129 | dlogz: 0.078 > 0.010]
- 6036it [00:10, 757.10it/s, batch: 0 | bound: 10 | nc: 1 | ncall: 24538 | eff(%): 24.107 | loglstar: -inf < -6.672 < inf | logz: -15.919 +/- 0.129 | dlogz: 0.062 > 0.010]
</pre>
- 6036it [00:10, 757.10it/s, batch: 0 | bound: 10 | nc: 1 | ncall: 24538 | eff(%): 24.107 | loglstar: -inf < -6.672 < inf | logz: -15.919 +/- 0.129 | dlogz: 0.062 > 0.010]
end{sphinxVerbatim}
6036it [00:10, 757.10it/s, batch: 0 | bound: 10 | nc: 1 | ncall: 24538 | eff(%): 24.107 | loglstar: -inf < -6.672 < inf | logz: -15.919 +/- 0.129 | dlogz: 0.062 > 0.010]
- 6136it [00:11, 601.03it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 24764 | eff(%): 24.288 | loglstar: -inf < -6.661 < inf | logz: -15.908 +/- 0.129 | dlogz: 0.051 > 0.010]
</pre>
- 6136it [00:11, 601.03it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 24764 | eff(%): 24.288 | loglstar: -inf < -6.661 < inf | logz: -15.908 +/- 0.129 | dlogz: 0.051 > 0.010]
end{sphinxVerbatim}
6136it [00:11, 601.03it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 24764 | eff(%): 24.288 | loglstar: -inf < -6.661 < inf | logz: -15.908 +/- 0.129 | dlogz: 0.051 > 0.010]
- 6281it [00:11, 756.44it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 24953 | eff(%): 24.677 | loglstar: -inf < -6.646 < inf | logz: -15.896 +/- 0.129 | dlogz: 0.038 > 0.010]
</pre>
- 6281it [00:11, 756.44it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 24953 | eff(%): 24.677 | loglstar: -inf < -6.646 < inf | logz: -15.896 +/- 0.129 | dlogz: 0.038 > 0.010]
end{sphinxVerbatim}
6281it [00:11, 756.44it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 24953 | eff(%): 24.677 | loglstar: -inf < -6.646 < inf | logz: -15.896 +/- 0.129 | dlogz: 0.038 > 0.010]
- 6440it [00:11, 929.33it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 25225 | eff(%): 25.034 | loglstar: -inf < -6.635 < inf | logz: -15.886 +/- 0.129 | dlogz: 0.027 > 0.010]
</pre>
- 6440it [00:11, 929.33it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 25225 | eff(%): 25.034 | loglstar: -inf < -6.635 < inf | logz: -15.886 +/- 0.129 | dlogz: 0.027 > 0.010]
end{sphinxVerbatim}
6440it [00:11, 929.33it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 25225 | eff(%): 25.034 | loglstar: -inf < -6.635 < inf | logz: -15.886 +/- 0.129 | dlogz: 0.027 > 0.010]
- 6556it [00:11, 943.56it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 25481 | eff(%): 25.234 | loglstar: -inf < -6.629 < inf | logz: -15.880 +/- 0.129 | dlogz: 0.022 > 0.010]
</pre>
- 6556it [00:11, 943.56it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 25481 | eff(%): 25.234 | loglstar: -inf < -6.629 < inf | logz: -15.880 +/- 0.129 | dlogz: 0.022 > 0.010]
end{sphinxVerbatim}
6556it [00:11, 943.56it/s, batch: 0 | bound: 11 | nc: 1 | ncall: 25481 | eff(%): 25.234 | loglstar: -inf < -6.629 < inf | logz: -15.880 +/- 0.129 | dlogz: 0.022 > 0.010]
- 6667it [00:11, 611.02it/s, batch: 0 | bound: 12 | nc: 1 | ncall: 25640 | eff(%): 25.505 | loglstar: -inf < -6.625 < inf | logz: -15.876 +/- 0.129 | dlogz: 0.017 > 0.010]
</pre>
- 6667it [00:11, 611.02it/s, batch: 0 | bound: 12 | nc: 1 | ncall: 25640 | eff(%): 25.505 | loglstar: -inf < -6.625 < inf | logz: -15.876 +/- 0.129 | dlogz: 0.017 > 0.010]
end{sphinxVerbatim}
6667it [00:11, 611.02it/s, batch: 0 | bound: 12 | nc: 1 | ncall: 25640 | eff(%): 25.505 | loglstar: -inf < -6.625 < inf | logz: -15.876 +/- 0.129 | dlogz: 0.017 > 0.010]
- 6805it [00:11, 745.15it/s, batch: 0 | bound: 12 | nc: 3 | ncall: 25863 | eff(%): 25.813 | loglstar: -inf < -6.621 < inf | logz: -15.872 +/- 0.129 | dlogz: 0.013 > 0.010]
</pre>
- 6805it [00:11, 745.15it/s, batch: 0 | bound: 12 | nc: 3 | ncall: 25863 | eff(%): 25.813 | loglstar: -inf < -6.621 < inf | logz: -15.872 +/- 0.129 | dlogz: 0.013 > 0.010]
end{sphinxVerbatim}
6805it [00:11, 745.15it/s, batch: 0 | bound: 12 | nc: 3 | ncall: 25863 | eff(%): 25.813 | loglstar: -inf < -6.621 < inf | logz: -15.872 +/- 0.129 | dlogz: 0.013 > 0.010]
- 6918it [00:11, 820.26it/s, batch: 0 | bound: 12 | nc: 4 | ncall: 26083 | eff(%): 26.024 | loglstar: -inf < -6.618 < inf | logz: -15.869 +/- 0.129 | dlogz: 0.010 > 0.010]
</pre>
- 6918it [00:11, 820.26it/s, batch: 0 | bound: 12 | nc: 4 | ncall: 26083 | eff(%): 26.024 | loglstar: -inf < -6.618 < inf | logz: -15.869 +/- 0.129 | dlogz: 0.010 > 0.010]
end{sphinxVerbatim}
6918it [00:11, 820.26it/s, batch: 0 | bound: 12 | nc: 4 | ncall: 26083 | eff(%): 26.024 | loglstar: -inf < -6.618 < inf | logz: -15.869 +/- 0.129 | dlogz: 0.010 > 0.010]
- 7157it [00:12, 1167.91it/s, batch: 0 | bound: 12 | nc: 1 | ncall: 26370 | eff(%): 26.855 | loglstar: -inf < -6.613 < inf | logz: -15.864 +/- 0.129 | dlogz: 0.006 > 0.010]
</pre>
- 7157it [00:12, 1167.91it/s, batch: 0 | bound: 12 | nc: 1 | ncall: 26370 | eff(%): 26.855 | loglstar: -inf < -6.613 < inf | logz: -15.864 +/- 0.129 | dlogz: 0.006 > 0.010]
end{sphinxVerbatim}
7157it [00:12, 1167.91it/s, batch: 0 | bound: 12 | nc: 1 | ncall: 26370 | eff(%): 26.855 | loglstar: -inf < -6.613 < inf | logz: -15.864 +/- 0.129 | dlogz: 0.006 > 0.010]
WARNING DeprecationWarning: This an old stopping function that will be removed in future releases
- 7438it [00:13, 433.94it/s, batch: 1 | bound: 0 | nc: 1 | ncall: 26651 | eff(%): 27.909 | loglstar: -8.346 < -7.850 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
</pre>
- 7438it [00:13, 433.94it/s, batch: 1 | bound: 0 | nc: 1 | ncall: 26651 | eff(%): 27.909 | loglstar: -8.346 < -7.850 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
end{sphinxVerbatim}
7438it [00:13, 433.94it/s, batch: 1 | bound: 0 | nc: 1 | ncall: 26651 | eff(%): 27.909 | loglstar: -8.346 < -7.850 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
- 7547it [00:13, 489.69it/s, batch: 1 | bound: 2 | nc: 1 | ncall: 26775 | eff(%): 27.637 | loglstar: -8.346 < -8.047 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
</pre>
- 7547it [00:13, 489.69it/s, batch: 1 | bound: 2 | nc: 1 | ncall: 26775 | eff(%): 27.637 | loglstar: -8.346 < -8.047 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
end{sphinxVerbatim}
7547it [00:13, 489.69it/s, batch: 1 | bound: 2 | nc: 1 | ncall: 26775 | eff(%): 27.637 | loglstar: -8.346 < -8.047 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
- 7655it [00:13, 552.73it/s, batch: 1 | bound: 2 | nc: 1 | ncall: 26956 | eff(%): 27.848 | loglstar: -8.346 < -7.750 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
</pre>
- 7655it [00:13, 552.73it/s, batch: 1 | bound: 2 | nc: 1 | ncall: 26956 | eff(%): 27.848 | loglstar: -8.346 < -7.750 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
end{sphinxVerbatim}
7655it [00:13, 552.73it/s, batch: 1 | bound: 2 | nc: 1 | ncall: 26956 | eff(%): 27.848 | loglstar: -8.346 < -7.750 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
- 7762it [00:13, 587.83it/s, batch: 1 | bound: 2 | nc: 4 | ncall: 27176 | eff(%): 28.013 | loglstar: -8.346 < -7.550 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
</pre>
- 7762it [00:13, 587.83it/s, batch: 1 | bound: 2 | nc: 4 | ncall: 27176 | eff(%): 28.013 | loglstar: -8.346 < -7.550 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
end{sphinxVerbatim}
7762it [00:13, 587.83it/s, batch: 1 | bound: 2 | nc: 4 | ncall: 27176 | eff(%): 28.013 | loglstar: -8.346 < -7.550 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
- 7859it [00:13, 571.61it/s, batch: 1 | bound: 2 | nc: 4 | ncall: 27386 | eff(%): 28.149 | loglstar: -8.346 < -7.370 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
</pre>
- 7859it [00:13, 571.61it/s, batch: 1 | bound: 2 | nc: 4 | ncall: 27386 | eff(%): 28.149 | loglstar: -8.346 < -7.370 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
end{sphinxVerbatim}
7859it [00:13, 571.61it/s, batch: 1 | bound: 2 | nc: 4 | ncall: 27386 | eff(%): 28.149 | loglstar: -8.346 < -7.370 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
- 7943it [00:14, 427.95it/s, batch: 1 | bound: 3 | nc: 3 | ncall: 27500 | eff(%): 28.334 | loglstar: -8.346 < -7.246 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
</pre>
- 7943it [00:14, 427.95it/s, batch: 1 | bound: 3 | nc: 3 | ncall: 27500 | eff(%): 28.334 | loglstar: -8.346 < -7.246 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
end{sphinxVerbatim}
7943it [00:14, 427.95it/s, batch: 1 | bound: 3 | nc: 3 | ncall: 27500 | eff(%): 28.334 | loglstar: -8.346 < -7.246 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
- 8217it [00:14, 750.73it/s, batch: 1 | bound: 3 | nc: 1 | ncall: 27850 | eff(%): 29.115 | loglstar: -8.346 < -6.958 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
</pre>
- 8217it [00:14, 750.73it/s, batch: 1 | bound: 3 | nc: 1 | ncall: 27850 | eff(%): 29.115 | loglstar: -8.346 < -6.958 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
end{sphinxVerbatim}
8217it [00:14, 750.73it/s, batch: 1 | bound: 3 | nc: 1 | ncall: 27850 | eff(%): 29.115 | loglstar: -8.346 < -6.958 < -7.113 | logz: -15.859 +/- 0.132 | stop: 1.406]
WARNING DeprecationWarning: This an old stopping function that will be removed in future releases
- 8568it [00:15, 421.98it/s, batch: 2 | bound: 0 | nc: 1 | ncall: 28223 | eff(%): 30.358 | loglstar: -8.865 < -6.918 < -8.345 | logz: -15.871 +/- 0.109 | stop: 1.147]
</pre>
- 8568it [00:15, 421.98it/s, batch: 2 | bound: 0 | nc: 1 | ncall: 28223 | eff(%): 30.358 | loglstar: -8.865 < -6.918 < -8.345 | logz: -15.871 +/- 0.109 | stop: 1.147]
end{sphinxVerbatim}
8568it [00:15, 421.98it/s, batch: 2 | bound: 0 | nc: 1 | ncall: 28223 | eff(%): 30.358 | loglstar: -8.865 < -6.918 < -8.345 | logz: -15.871 +/- 0.109 | stop: 1.147]
- 8787it [00:15, 557.24it/s, batch: 2 | bound: 2 | nc: 1 | ncall: 28479 | eff(%): 30.365 | loglstar: -8.865 < -7.967 < -8.345 | logz: -15.871 +/- 0.109 | stop: 1.147]
</pre>
- 8787it [00:15, 557.24it/s, batch: 2 | bound: 2 | nc: 1 | ncall: 28479 | eff(%): 30.365 | loglstar: -8.865 < -7.967 < -8.345 | logz: -15.871 +/- 0.109 | stop: 1.147]
end{sphinxVerbatim}
8787it [00:15, 557.24it/s, batch: 2 | bound: 2 | nc: 1 | ncall: 28479 | eff(%): 30.365 | loglstar: -8.865 < -7.967 < -8.345 | logz: -15.871 +/- 0.109 | stop: 1.147]
WARNING DeprecationWarning: This an old stopping function that will be removed in future releases
- 9191it [00:16, 416.98it/s, batch: 3 | bound: 0 | nc: 1 | ncall: 28938 | eff(%): 31.761 | loglstar: -9.259 < -8.161 < -8.862 | logz: -15.860 +/- 0.101 | stop: 1.033]
</pre>
- 9191it [00:16, 416.98it/s, batch: 3 | bound: 0 | nc: 1 | ncall: 28938 | eff(%): 31.761 | loglstar: -9.259 < -8.161 < -8.862 | logz: -15.860 +/- 0.101 | stop: 1.033]
end{sphinxVerbatim}
9191it [00:16, 416.98it/s, batch: 3 | bound: 0 | nc: 1 | ncall: 28938 | eff(%): 31.761 | loglstar: -9.259 < -8.161 < -8.862 | logz: -15.860 +/- 0.101 | stop: 1.033]
- 9338it [00:16, 481.35it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29105 | eff(%): 31.600 | loglstar: -9.259 < -8.532 < -8.862 | logz: -15.860 +/- 0.101 | stop: 1.033]
</pre>
- 9338it [00:16, 481.35it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29105 | eff(%): 31.600 | loglstar: -9.259 < -8.532 < -8.862 | logz: -15.860 +/- 0.101 | stop: 1.033]
end{sphinxVerbatim}
9338it [00:16, 481.35it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29105 | eff(%): 31.600 | loglstar: -9.259 < -8.532 < -8.862 | logz: -15.860 +/- 0.101 | stop: 1.033]
- 9717it [00:17, 747.30it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29493 | eff(%): 32.882 | loglstar: -9.259 < -6.862 < -8.862 | logz: -15.860 +/- 0.101 | stop: 1.033]
</pre>
- 9717it [00:17, 747.30it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29493 | eff(%): 32.882 | loglstar: -9.259 < -6.862 < -8.862 | logz: -15.860 +/- 0.101 | stop: 1.033]
end{sphinxVerbatim}
9717it [00:17, 747.30it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29493 | eff(%): 32.882 | loglstar: -9.259 < -6.862 < -8.862 | logz: -15.860 +/- 0.101 | stop: 1.033]
WARNING DeprecationWarning: This an old stopping function that will be removed in future releases
- 9772it [00:18, 540.54it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29551 | eff(%): 33.068 | loglstar: -9.259 < -6.608 < -8.862 | logz: -15.860 +/- 0.101 | stop: 0.998]
</pre>
- 9772it [00:18, 540.54it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29551 | eff(%): 33.068 | loglstar: -9.259 < -6.608 < -8.862 | logz: -15.860 +/- 0.101 | stop: 0.998]
end{sphinxVerbatim}
9772it [00:18, 540.54it/s, batch: 3 | bound: 2 | nc: 1 | ncall: 29551 | eff(%): 33.068 | loglstar: -9.259 < -6.608 < -8.862 | logz: -15.860 +/- 0.101 | stop: 0.998]
22:19:19 INFO fit restored to maximum of posterior sampler_base.py:178
INFO fit restored to maximum of posterior sampler_base.py:178
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | (9.91 +/- 0.16) x 10^-1 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (9.99 +/- 0.05) x 10^-2 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -6.59844 |
| total | -6.59844 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 17.902762 |
| BIC | 19.188344 |
| DIC | 16.982383 |
| PDIC | 1.892672 |
| log(Z) | -6.889652 |
[8]:
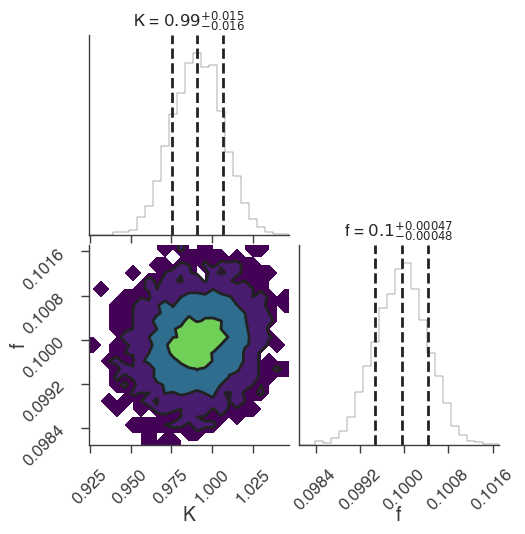
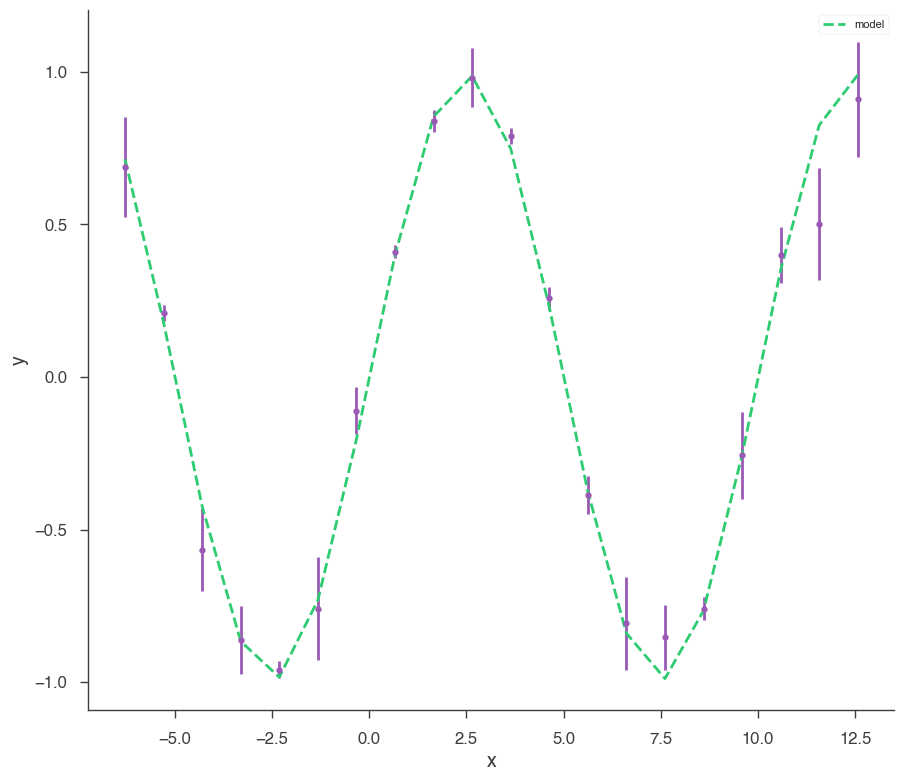
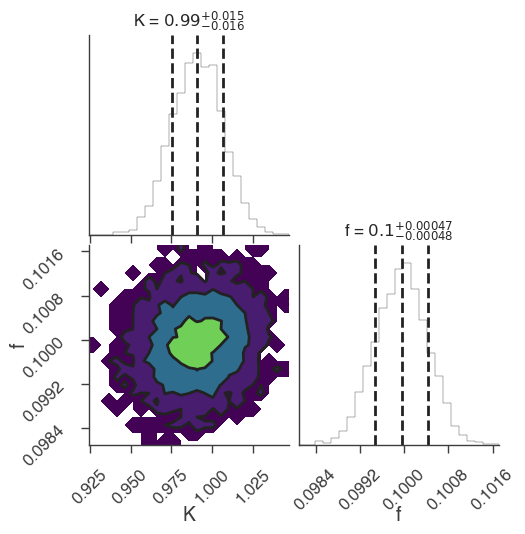
zeus
[9]:
bayes_analysis.set_sampler("zeus")
bayes_analysis.sampler.setup(n_walkers=20, n_iterations=500)
bayes_analysis.sample()
xyl.plot()
bayes_analysis.results.corner_plot()
22:19:21 INFO sampler set to zeus bayesian_analysis.py:202
WARNING:root:The sampler class has been deprecated. Please use the new EnsembleSampler class.
The run method has been deprecated and it will be removed. Please use the new run_mcmc method.
Initialising ensemble of 20 walkers...
- Sampling progress0%| | 0/625 [00:00<?, ?it/s]
</pre>
- Sampling progress0%| | 0/625 [00:00<?, ?it/s]
end{sphinxVerbatim}
Sampling progress : 0%| | 0/625 [00:00<?, ?it/s]
- Sampling progress1%| | 5/625 [00:00<00:16, 37.05it/s]
</pre>
- Sampling progress1%| | 5/625 [00:00<00:16, 37.05it/s]
end{sphinxVerbatim}
Sampling progress : 1%| | 5/625 [00:00<00:16, 37.05it/s]
- Sampling progress1%|▏ | 9/625 [00:00<00:20, 30.64it/s]
</pre>
- Sampling progress1%|▏ | 9/625 [00:00<00:20, 30.64it/s]
end{sphinxVerbatim}
Sampling progress : 1%|▏ | 9/625 [00:00<00:20, 30.64it/s]
- Sampling progress2%|▏ | 13/625 [00:00<00:20, 29.41it/s]
</pre>
- Sampling progress2%|▏ | 13/625 [00:00<00:20, 29.41it/s]
end{sphinxVerbatim}
Sampling progress : 2%|▏ | 13/625 [00:00<00:20, 29.41it/s]
- Sampling progress3%|▎ | 18/625 [00:00<00:17, 33.91it/s]
</pre>
- Sampling progress3%|▎ | 18/625 [00:00<00:17, 33.91it/s]
end{sphinxVerbatim}
Sampling progress : 3%|▎ | 18/625 [00:00<00:17, 33.91it/s]
- Sampling progress4%|▎ | 22/625 [00:00<00:18, 33.45it/s]
</pre>
- Sampling progress4%|▎ | 22/625 [00:00<00:18, 33.45it/s]
end{sphinxVerbatim}
Sampling progress : 4%|▎ | 22/625 [00:00<00:18, 33.45it/s]
- Sampling progress4%|▍ | 26/625 [00:00<00:18, 32.46it/s]
</pre>
- Sampling progress4%|▍ | 26/625 [00:00<00:18, 32.46it/s]
end{sphinxVerbatim}
Sampling progress : 4%|▍ | 26/625 [00:00<00:18, 32.46it/s]
- Sampling progress5%|▍ | 30/625 [00:00<00:18, 32.39it/s]
</pre>
- Sampling progress5%|▍ | 30/625 [00:00<00:18, 32.39it/s]
end{sphinxVerbatim}
Sampling progress : 5%|▍ | 30/625 [00:00<00:18, 32.39it/s]
- Sampling progress5%|▌ | 34/625 [00:01<00:18, 32.38it/s]
</pre>
- Sampling progress5%|▌ | 34/625 [00:01<00:18, 32.38it/s]
end{sphinxVerbatim}
Sampling progress : 5%|▌ | 34/625 [00:01<00:18, 32.38it/s]
- Sampling progress6%|▌ | 39/625 [00:01<00:16, 35.46it/s]
</pre>
- Sampling progress6%|▌ | 39/625 [00:01<00:16, 35.46it/s]
end{sphinxVerbatim}
Sampling progress : 6%|▌ | 39/625 [00:01<00:16, 35.46it/s]
- Sampling progress7%|▋ | 45/625 [00:01<00:14, 40.21it/s]
</pre>
- Sampling progress7%|▋ | 45/625 [00:01<00:14, 40.21it/s]
end{sphinxVerbatim}
Sampling progress : 7%|▋ | 45/625 [00:01<00:14, 40.21it/s]
- Sampling progress8%|▊ | 50/625 [00:01<00:15, 37.33it/s]
</pre>
- Sampling progress8%|▊ | 50/625 [00:01<00:15, 37.33it/s]
end{sphinxVerbatim}
Sampling progress : 8%|▊ | 50/625 [00:01<00:15, 37.33it/s]
- Sampling progress9%|▊ | 54/625 [00:01<00:20, 28.29it/s]
</pre>
- Sampling progress9%|▊ | 54/625 [00:01<00:20, 28.29it/s]
end{sphinxVerbatim}
Sampling progress : 9%|▊ | 54/625 [00:01<00:20, 28.29it/s]
- Sampling progress9%|▉ | 58/625 [00:01<00:19, 28.80it/s]
</pre>
- Sampling progress9%|▉ | 58/625 [00:01<00:19, 28.80it/s]
end{sphinxVerbatim}
Sampling progress : 9%|▉ | 58/625 [00:01<00:19, 28.80it/s]
- Sampling progress10%|▉ | 62/625 [00:01<00:18, 30.55it/s]
</pre>
- Sampling progress10%|▉ | 62/625 [00:01<00:18, 30.55it/s]
end{sphinxVerbatim}
Sampling progress : 10%|▉ | 62/625 [00:01<00:18, 30.55it/s]
- Sampling progress11%|█ | 67/625 [00:02<00:17, 32.22it/s]
</pre>
- Sampling progress11%|█ | 67/625 [00:02<00:17, 32.22it/s]
end{sphinxVerbatim}
Sampling progress : 11%|█ | 67/625 [00:02<00:17, 32.22it/s]
- Sampling progress11%|█▏ | 71/625 [00:02<00:16, 33.59it/s]
</pre>
- Sampling progress11%|█▏ | 71/625 [00:02<00:16, 33.59it/s]
end{sphinxVerbatim}
Sampling progress : 11%|█▏ | 71/625 [00:02<00:16, 33.59it/s]
- Sampling progress12%|█▏ | 75/625 [00:02<00:16, 32.53it/s]
</pre>
- Sampling progress12%|█▏ | 75/625 [00:02<00:16, 32.53it/s]
end{sphinxVerbatim}
Sampling progress : 12%|█▏ | 75/625 [00:02<00:16, 32.53it/s]
- Sampling progress13%|█▎ | 79/625 [00:02<00:16, 33.35it/s]
</pre>
- Sampling progress13%|█▎ | 79/625 [00:02<00:16, 33.35it/s]
end{sphinxVerbatim}
Sampling progress : 13%|█▎ | 79/625 [00:02<00:16, 33.35it/s]
- Sampling progress13%|█▎ | 83/625 [00:02<00:15, 34.59it/s]
</pre>
- Sampling progress13%|█▎ | 83/625 [00:02<00:15, 34.59it/s]
end{sphinxVerbatim}
Sampling progress : 13%|█▎ | 83/625 [00:02<00:15, 34.59it/s]
- Sampling progress14%|█▍ | 88/625 [00:02<00:15, 35.80it/s]
</pre>
- Sampling progress14%|█▍ | 88/625 [00:02<00:15, 35.80it/s]
end{sphinxVerbatim}
Sampling progress : 14%|█▍ | 88/625 [00:02<00:15, 35.80it/s]
- Sampling progress15%|█▍ | 93/625 [00:02<00:14, 37.55it/s]
</pre>
- Sampling progress15%|█▍ | 93/625 [00:02<00:14, 37.55it/s]
end{sphinxVerbatim}
Sampling progress : 15%|█▍ | 93/625 [00:02<00:14, 37.55it/s]
- Sampling progress16%|█▌ | 97/625 [00:02<00:18, 28.14it/s]
</pre>
- Sampling progress16%|█▌ | 97/625 [00:02<00:18, 28.14it/s]
end{sphinxVerbatim}
Sampling progress : 16%|█▌ | 97/625 [00:02<00:18, 28.14it/s]
- Sampling progress16%|█▋ | 103/625 [00:03<00:15, 34.28it/s]
</pre>
- Sampling progress16%|█▋ | 103/625 [00:03<00:15, 34.28it/s]
end{sphinxVerbatim}
Sampling progress : 16%|█▋ | 103/625 [00:03<00:15, 34.28it/s]
- Sampling progress17%|█▋ | 109/625 [00:03<00:13, 39.61it/s]
</pre>
- Sampling progress17%|█▋ | 109/625 [00:03<00:13, 39.61it/s]
end{sphinxVerbatim}
Sampling progress : 17%|█▋ | 109/625 [00:03<00:13, 39.61it/s]
- Sampling progress18%|█▊ | 114/625 [00:03<00:16, 31.64it/s]
</pre>
- Sampling progress18%|█▊ | 114/625 [00:03<00:16, 31.64it/s]
end{sphinxVerbatim}
Sampling progress : 18%|█▊ | 114/625 [00:03<00:16, 31.64it/s]
- Sampling progress19%|█▉ | 118/625 [00:03<00:15, 32.07it/s]
</pre>
- Sampling progress19%|█▉ | 118/625 [00:03<00:15, 32.07it/s]
end{sphinxVerbatim}
Sampling progress : 19%|█▉ | 118/625 [00:03<00:15, 32.07it/s]
- Sampling progress20%|█▉ | 122/625 [00:03<00:15, 33.14it/s]
</pre>
- Sampling progress20%|█▉ | 122/625 [00:03<00:15, 33.14it/s]
end{sphinxVerbatim}
Sampling progress : 20%|█▉ | 122/625 [00:03<00:15, 33.14it/s]
- Sampling progress20%|██ | 127/625 [00:03<00:14, 35.35it/s]
</pre>
- Sampling progress20%|██ | 127/625 [00:03<00:14, 35.35it/s]
end{sphinxVerbatim}
Sampling progress : 20%|██ | 127/625 [00:03<00:14, 35.35it/s]
- Sampling progress21%|██ | 131/625 [00:03<00:14, 34.74it/s]
</pre>
- Sampling progress21%|██ | 131/625 [00:03<00:14, 34.74it/s]
end{sphinxVerbatim}
Sampling progress : 21%|██ | 131/625 [00:03<00:14, 34.74it/s]
- Sampling progress22%|██▏ | 136/625 [00:04<00:13, 36.68it/s]
</pre>
- Sampling progress22%|██▏ | 136/625 [00:04<00:13, 36.68it/s]
end{sphinxVerbatim}
Sampling progress : 22%|██▏ | 136/625 [00:04<00:13, 36.68it/s]
- Sampling progress22%|██▏ | 140/625 [00:04<00:13, 35.81it/s]
</pre>
- Sampling progress22%|██▏ | 140/625 [00:04<00:13, 35.81it/s]
end{sphinxVerbatim}
Sampling progress : 22%|██▏ | 140/625 [00:04<00:13, 35.81it/s]
- Sampling progress23%|██▎ | 144/625 [00:04<00:13, 35.56it/s]
</pre>
- Sampling progress23%|██▎ | 144/625 [00:04<00:13, 35.56it/s]
end{sphinxVerbatim}
Sampling progress : 23%|██▎ | 144/625 [00:04<00:13, 35.56it/s]
- Sampling progress24%|██▍ | 149/625 [00:04<00:12, 39.27it/s]
</pre>
- Sampling progress24%|██▍ | 149/625 [00:04<00:12, 39.27it/s]
end{sphinxVerbatim}
Sampling progress : 24%|██▍ | 149/625 [00:04<00:12, 39.27it/s]
- Sampling progress25%|██▍ | 154/625 [00:04<00:12, 37.65it/s]
</pre>
- Sampling progress25%|██▍ | 154/625 [00:04<00:12, 37.65it/s]
end{sphinxVerbatim}
Sampling progress : 25%|██▍ | 154/625 [00:04<00:12, 37.65it/s]
- Sampling progress25%|██▌ | 158/625 [00:04<00:15, 31.00it/s]
</pre>
- Sampling progress25%|██▌ | 158/625 [00:04<00:15, 31.00it/s]
end{sphinxVerbatim}
Sampling progress : 25%|██▌ | 158/625 [00:04<00:15, 31.00it/s]
- Sampling progress26%|██▌ | 162/625 [00:04<00:14, 32.23it/s]
</pre>
- Sampling progress26%|██▌ | 162/625 [00:04<00:14, 32.23it/s]
end{sphinxVerbatim}
Sampling progress : 26%|██▌ | 162/625 [00:04<00:14, 32.23it/s]
- Sampling progress27%|██▋ | 167/625 [00:04<00:13, 34.67it/s]
</pre>
- Sampling progress27%|██▋ | 167/625 [00:04<00:13, 34.67it/s]
end{sphinxVerbatim}
Sampling progress : 27%|██▋ | 167/625 [00:04<00:13, 34.67it/s]
- Sampling progress27%|██▋ | 171/625 [00:05<00:12, 35.66it/s]
</pre>
- Sampling progress27%|██▋ | 171/625 [00:05<00:12, 35.66it/s]
end{sphinxVerbatim}
Sampling progress : 27%|██▋ | 171/625 [00:05<00:12, 35.66it/s]
- Sampling progress28%|██▊ | 175/625 [00:05<00:12, 35.97it/s]
</pre>
- Sampling progress28%|██▊ | 175/625 [00:05<00:12, 35.97it/s]
end{sphinxVerbatim}
Sampling progress : 28%|██▊ | 175/625 [00:05<00:12, 35.97it/s]
- Sampling progress29%|██▉ | 180/625 [00:05<00:11, 37.69it/s]
</pre>
- Sampling progress29%|██▉ | 180/625 [00:05<00:11, 37.69it/s]
end{sphinxVerbatim}
Sampling progress : 29%|██▉ | 180/625 [00:05<00:11, 37.69it/s]
- Sampling progress30%|██▉ | 185/625 [00:05<00:11, 39.95it/s]
</pre>
- Sampling progress30%|██▉ | 185/625 [00:05<00:11, 39.95it/s]
end{sphinxVerbatim}
Sampling progress : 30%|██▉ | 185/625 [00:05<00:11, 39.95it/s]
- Sampling progress30%|███ | 190/625 [00:05<00:14, 30.29it/s]
</pre>
- Sampling progress30%|███ | 190/625 [00:05<00:14, 30.29it/s]
end{sphinxVerbatim}
Sampling progress : 30%|███ | 190/625 [00:05<00:14, 30.29it/s]
- Sampling progress31%|███ | 195/625 [00:05<00:13, 32.85it/s]
</pre>
- Sampling progress31%|███ | 195/625 [00:05<00:13, 32.85it/s]
end{sphinxVerbatim}
Sampling progress : 31%|███ | 195/625 [00:05<00:13, 32.85it/s]
- Sampling progress32%|███▏ | 200/625 [00:05<00:11, 35.55it/s]
</pre>
- Sampling progress32%|███▏ | 200/625 [00:05<00:11, 35.55it/s]
end{sphinxVerbatim}
Sampling progress : 32%|███▏ | 200/625 [00:05<00:11, 35.55it/s]
- Sampling progress33%|███▎ | 207/625 [00:05<00:09, 42.17it/s]
</pre>
- Sampling progress33%|███▎ | 207/625 [00:05<00:09, 42.17it/s]
end{sphinxVerbatim}
Sampling progress : 33%|███▎ | 207/625 [00:05<00:09, 42.17it/s]
- Sampling progress34%|███▍ | 213/625 [00:06<00:09, 45.66it/s]
</pre>
- Sampling progress34%|███▍ | 213/625 [00:06<00:09, 45.66it/s]
end{sphinxVerbatim}
Sampling progress : 34%|███▍ | 213/625 [00:06<00:09, 45.66it/s]
- Sampling progress35%|███▍ | 218/625 [00:06<00:09, 44.14it/s]
</pre>
- Sampling progress35%|███▍ | 218/625 [00:06<00:09, 44.14it/s]
end{sphinxVerbatim}
Sampling progress : 35%|███▍ | 218/625 [00:06<00:09, 44.14it/s]
- Sampling progress36%|███▌ | 223/625 [00:06<00:09, 43.07it/s]
</pre>
- Sampling progress36%|███▌ | 223/625 [00:06<00:09, 43.07it/s]
end{sphinxVerbatim}
Sampling progress : 36%|███▌ | 223/625 [00:06<00:09, 43.07it/s]
- Sampling progress37%|███▋ | 229/625 [00:06<00:08, 46.58it/s]
</pre>
- Sampling progress37%|███▋ | 229/625 [00:06<00:08, 46.58it/s]
end{sphinxVerbatim}
Sampling progress : 37%|███▋ | 229/625 [00:06<00:08, 46.58it/s]
- Sampling progress37%|███▋ | 234/625 [00:06<00:08, 46.52it/s]
</pre>
- Sampling progress37%|███▋ | 234/625 [00:06<00:08, 46.52it/s]
end{sphinxVerbatim}
Sampling progress : 37%|███▋ | 234/625 [00:06<00:08, 46.52it/s]
- Sampling progress38%|███▊ | 240/625 [00:06<00:08, 47.80it/s]
</pre>
- Sampling progress38%|███▊ | 240/625 [00:06<00:08, 47.80it/s]
end{sphinxVerbatim}
Sampling progress : 38%|███▊ | 240/625 [00:06<00:08, 47.80it/s]
- Sampling progress39%|███▉ | 245/625 [00:06<00:07, 47.97it/s]
</pre>
- Sampling progress39%|███▉ | 245/625 [00:06<00:07, 47.97it/s]
end{sphinxVerbatim}
Sampling progress : 39%|███▉ | 245/625 [00:06<00:07, 47.97it/s]
- Sampling progress40%|████ | 251/625 [00:06<00:07, 49.20it/s]
</pre>
- Sampling progress40%|████ | 251/625 [00:06<00:07, 49.20it/s]
end{sphinxVerbatim}
Sampling progress : 40%|████ | 251/625 [00:06<00:07, 49.20it/s]
- Sampling progress41%|████ | 257/625 [00:07<00:07, 51.00it/s]
</pre>
- Sampling progress41%|████ | 257/625 [00:07<00:07, 51.00it/s]
end{sphinxVerbatim}
Sampling progress : 41%|████ | 257/625 [00:07<00:07, 51.00it/s]
- Sampling progress42%|████▏ | 263/625 [00:07<00:06, 52.25it/s]
</pre>
- Sampling progress42%|████▏ | 263/625 [00:07<00:06, 52.25it/s]
end{sphinxVerbatim}
Sampling progress : 42%|████▏ | 263/625 [00:07<00:06, 52.25it/s]
- Sampling progress43%|████▎ | 269/625 [00:07<00:06, 54.02it/s]
</pre>
- Sampling progress43%|████▎ | 269/625 [00:07<00:06, 54.02it/s]
end{sphinxVerbatim}
Sampling progress : 43%|████▎ | 269/625 [00:07<00:06, 54.02it/s]
- Sampling progress44%|████▍ | 275/625 [00:07<00:06, 50.64it/s]
</pre>
- Sampling progress44%|████▍ | 275/625 [00:07<00:06, 50.64it/s]
end{sphinxVerbatim}
Sampling progress : 44%|████▍ | 275/625 [00:07<00:06, 50.64it/s]
- Sampling progress45%|████▍ | 281/625 [00:07<00:07, 48.72it/s]
</pre>
- Sampling progress45%|████▍ | 281/625 [00:07<00:07, 48.72it/s]
end{sphinxVerbatim}
Sampling progress : 45%|████▍ | 281/625 [00:07<00:07, 48.72it/s]
- Sampling progress46%|████▌ | 286/625 [00:07<00:07, 46.47it/s]
</pre>
- Sampling progress46%|████▌ | 286/625 [00:07<00:07, 46.47it/s]
end{sphinxVerbatim}
Sampling progress : 46%|████▌ | 286/625 [00:07<00:07, 46.47it/s]
- Sampling progress47%|████▋ | 291/625 [00:07<00:09, 34.72it/s]
</pre>
- Sampling progress47%|████▋ | 291/625 [00:07<00:09, 34.72it/s]
end{sphinxVerbatim}
Sampling progress : 47%|████▋ | 291/625 [00:07<00:09, 34.72it/s]
- Sampling progress47%|████▋ | 296/625 [00:08<00:11, 28.40it/s]
</pre>
- Sampling progress47%|████▋ | 296/625 [00:08<00:11, 28.40it/s]
end{sphinxVerbatim}
Sampling progress : 47%|████▋ | 296/625 [00:08<00:11, 28.40it/s]
- Sampling progress48%|████▊ | 300/625 [00:08<00:12, 25.31it/s]
</pre>
- Sampling progress48%|████▊ | 300/625 [00:08<00:12, 25.31it/s]
end{sphinxVerbatim}
Sampling progress : 48%|████▊ | 300/625 [00:08<00:12, 25.31it/s]
- Sampling progress49%|████▊ | 304/625 [00:08<00:11, 27.84it/s]
</pre>
- Sampling progress49%|████▊ | 304/625 [00:08<00:11, 27.84it/s]
end{sphinxVerbatim}
Sampling progress : 49%|████▊ | 304/625 [00:08<00:11, 27.84it/s]
- Sampling progress49%|████▉ | 308/625 [00:08<00:11, 28.79it/s]
</pre>
- Sampling progress49%|████▉ | 308/625 [00:08<00:11, 28.79it/s]
end{sphinxVerbatim}
Sampling progress : 49%|████▉ | 308/625 [00:08<00:11, 28.79it/s]
- Sampling progress50%|████▉ | 312/625 [00:08<00:12, 25.12it/s]
</pre>
- Sampling progress50%|████▉ | 312/625 [00:08<00:12, 25.12it/s]
end{sphinxVerbatim}
Sampling progress : 50%|████▉ | 312/625 [00:08<00:12, 25.12it/s]
- Sampling progress51%|█████ | 318/625 [00:08<00:09, 31.72it/s]
</pre>
- Sampling progress51%|█████ | 318/625 [00:08<00:09, 31.72it/s]
end{sphinxVerbatim}
Sampling progress : 51%|█████ | 318/625 [00:08<00:09, 31.72it/s]
- Sampling progress52%|█████▏ | 323/625 [00:08<00:08, 35.13it/s]
</pre>
- Sampling progress52%|█████▏ | 323/625 [00:08<00:08, 35.13it/s]
end{sphinxVerbatim}
Sampling progress : 52%|█████▏ | 323/625 [00:08<00:08, 35.13it/s]
- Sampling progress52%|█████▏ | 327/625 [00:09<00:10, 28.45it/s]
</pre>
- Sampling progress52%|█████▏ | 327/625 [00:09<00:10, 28.45it/s]
end{sphinxVerbatim}
Sampling progress : 52%|█████▏ | 327/625 [00:09<00:10, 28.45it/s]
- Sampling progress53%|█████▎ | 333/625 [00:09<00:13, 22.02it/s]
</pre>
- Sampling progress53%|█████▎ | 333/625 [00:09<00:13, 22.02it/s]
end{sphinxVerbatim}
Sampling progress : 53%|█████▎ | 333/625 [00:09<00:13, 22.02it/s]
- Sampling progress54%|█████▍ | 338/625 [00:09<00:12, 23.88it/s]
</pre>
- Sampling progress54%|█████▍ | 338/625 [00:09<00:12, 23.88it/s]
end{sphinxVerbatim}
Sampling progress : 54%|█████▍ | 338/625 [00:09<00:12, 23.88it/s]
- Sampling progress55%|█████▍ | 341/625 [00:09<00:13, 20.93it/s]
</pre>
- Sampling progress55%|█████▍ | 341/625 [00:09<00:13, 20.93it/s]
end{sphinxVerbatim}
Sampling progress : 55%|█████▍ | 341/625 [00:09<00:13, 20.93it/s]
- Sampling progress55%|█████▌ | 345/625 [00:10<00:11, 23.59it/s]
</pre>
- Sampling progress55%|█████▌ | 345/625 [00:10<00:11, 23.59it/s]
end{sphinxVerbatim}
Sampling progress : 55%|█████▌ | 345/625 [00:10<00:11, 23.59it/s]
- Sampling progress56%|█████▌ | 349/625 [00:10<00:12, 21.43it/s]
</pre>
- Sampling progress56%|█████▌ | 349/625 [00:10<00:12, 21.43it/s]
end{sphinxVerbatim}
Sampling progress : 56%|█████▌ | 349/625 [00:10<00:12, 21.43it/s]
- Sampling progress56%|█████▋ | 352/625 [00:10<00:12, 21.92it/s]
</pre>
- Sampling progress56%|█████▋ | 352/625 [00:10<00:12, 21.92it/s]
end{sphinxVerbatim}
Sampling progress : 56%|█████▋ | 352/625 [00:10<00:12, 21.92it/s]
- Sampling progress57%|█████▋ | 355/625 [00:10<00:11, 23.41it/s]
</pre>
- Sampling progress57%|█████▋ | 355/625 [00:10<00:11, 23.41it/s]
end{sphinxVerbatim}
Sampling progress : 57%|█████▋ | 355/625 [00:10<00:11, 23.41it/s]
- Sampling progress57%|█████▋ | 359/625 [00:10<00:09, 26.71it/s]
</pre>
- Sampling progress57%|█████▋ | 359/625 [00:10<00:09, 26.71it/s]
end{sphinxVerbatim}
Sampling progress : 57%|█████▋ | 359/625 [00:10<00:09, 26.71it/s]
- Sampling progress58%|█████▊ | 362/625 [00:10<00:09, 27.21it/s]
</pre>
- Sampling progress58%|█████▊ | 362/625 [00:10<00:09, 27.21it/s]
end{sphinxVerbatim}
Sampling progress : 58%|█████▊ | 362/625 [00:10<00:09, 27.21it/s]
- Sampling progress59%|█████▊ | 366/625 [00:10<00:08, 29.81it/s]
</pre>
- Sampling progress59%|█████▊ | 366/625 [00:10<00:08, 29.81it/s]
end{sphinxVerbatim}
Sampling progress : 59%|█████▊ | 366/625 [00:10<00:08, 29.81it/s]
- Sampling progress59%|█████▉ | 370/625 [00:10<00:07, 31.93it/s]
</pre>
- Sampling progress59%|█████▉ | 370/625 [00:10<00:07, 31.93it/s]
end{sphinxVerbatim}
Sampling progress : 59%|█████▉ | 370/625 [00:10<00:07, 31.93it/s]
- Sampling progress60%|█████▉ | 374/625 [00:11<00:08, 31.07it/s]
</pre>
- Sampling progress60%|█████▉ | 374/625 [00:11<00:08, 31.07it/s]
end{sphinxVerbatim}
Sampling progress : 60%|█████▉ | 374/625 [00:11<00:08, 31.07it/s]
- Sampling progress60%|██████ | 378/625 [00:11<00:08, 30.09it/s]
</pre>
- Sampling progress60%|██████ | 378/625 [00:11<00:08, 30.09it/s]
end{sphinxVerbatim}
Sampling progress : 60%|██████ | 378/625 [00:11<00:08, 30.09it/s]
- Sampling progress61%|██████ | 382/625 [00:11<00:07, 31.62it/s]
</pre>
- Sampling progress61%|██████ | 382/625 [00:11<00:07, 31.62it/s]
end{sphinxVerbatim}
Sampling progress : 61%|██████ | 382/625 [00:11<00:07, 31.62it/s]
- Sampling progress62%|██████▏ | 387/625 [00:11<00:06, 34.76it/s]
</pre>
- Sampling progress62%|██████▏ | 387/625 [00:11<00:06, 34.76it/s]
end{sphinxVerbatim}
Sampling progress : 62%|██████▏ | 387/625 [00:11<00:06, 34.76it/s]
- Sampling progress63%|██████▎ | 392/625 [00:11<00:06, 38.21it/s]
</pre>
- Sampling progress63%|██████▎ | 392/625 [00:11<00:06, 38.21it/s]
end{sphinxVerbatim}
Sampling progress : 63%|██████▎ | 392/625 [00:11<00:06, 38.21it/s]
- Sampling progress64%|██████▎ | 397/625 [00:11<00:05, 39.10it/s]
</pre>
- Sampling progress64%|██████▎ | 397/625 [00:11<00:05, 39.10it/s]
end{sphinxVerbatim}
Sampling progress : 64%|██████▎ | 397/625 [00:11<00:05, 39.10it/s]
- Sampling progress64%|██████▍ | 401/625 [00:12<00:10, 21.43it/s]
</pre>
- Sampling progress64%|██████▍ | 401/625 [00:12<00:10, 21.43it/s]
end{sphinxVerbatim}
Sampling progress : 64%|██████▍ | 401/625 [00:12<00:10, 21.43it/s]
- Sampling progress65%|██████▍ | 406/625 [00:12<00:08, 25.48it/s]
</pre>
- Sampling progress65%|██████▍ | 406/625 [00:12<00:08, 25.48it/s]
end{sphinxVerbatim}
Sampling progress : 65%|██████▍ | 406/625 [00:12<00:08, 25.48it/s]
- Sampling progress66%|██████▌ | 411/625 [00:12<00:07, 29.60it/s]
</pre>
- Sampling progress66%|██████▌ | 411/625 [00:12<00:07, 29.60it/s]
end{sphinxVerbatim}
Sampling progress : 66%|██████▌ | 411/625 [00:12<00:07, 29.60it/s]
- Sampling progress67%|██████▋ | 416/625 [00:12<00:06, 32.79it/s]
</pre>
- Sampling progress67%|██████▋ | 416/625 [00:12<00:06, 32.79it/s]
end{sphinxVerbatim}
Sampling progress : 67%|██████▋ | 416/625 [00:12<00:06, 32.79it/s]
- Sampling progress67%|██████▋ | 421/625 [00:12<00:05, 35.08it/s]
</pre>
- Sampling progress67%|██████▋ | 421/625 [00:12<00:05, 35.08it/s]
end{sphinxVerbatim}
Sampling progress : 67%|██████▋ | 421/625 [00:12<00:05, 35.08it/s]
- Sampling progress68%|██████▊ | 425/625 [00:12<00:05, 33.34it/s]
</pre>
- Sampling progress68%|██████▊ | 425/625 [00:12<00:05, 33.34it/s]
end{sphinxVerbatim}
Sampling progress : 68%|██████▊ | 425/625 [00:12<00:05, 33.34it/s]
- Sampling progress69%|██████▊ | 429/625 [00:12<00:05, 33.10it/s]
</pre>
- Sampling progress69%|██████▊ | 429/625 [00:12<00:05, 33.10it/s]
end{sphinxVerbatim}
Sampling progress : 69%|██████▊ | 429/625 [00:12<00:05, 33.10it/s]
- Sampling progress69%|██████▉ | 433/625 [00:12<00:06, 31.51it/s]
</pre>
- Sampling progress69%|██████▉ | 433/625 [00:12<00:06, 31.51it/s]
end{sphinxVerbatim}
Sampling progress : 69%|██████▉ | 433/625 [00:12<00:06, 31.51it/s]
- Sampling progress70%|██████▉ | 437/625 [00:13<00:06, 29.50it/s]
</pre>
- Sampling progress70%|██████▉ | 437/625 [00:13<00:06, 29.50it/s]
end{sphinxVerbatim}
Sampling progress : 70%|██████▉ | 437/625 [00:13<00:06, 29.50it/s]
- Sampling progress71%|███████ | 443/625 [00:13<00:05, 36.27it/s]
</pre>
- Sampling progress71%|███████ | 443/625 [00:13<00:05, 36.27it/s]
end{sphinxVerbatim}
Sampling progress : 71%|███████ | 443/625 [00:13<00:05, 36.27it/s]
- Sampling progress72%|███████▏ | 449/625 [00:13<00:04, 40.21it/s]
</pre>
- Sampling progress72%|███████▏ | 449/625 [00:13<00:04, 40.21it/s]
end{sphinxVerbatim}
Sampling progress : 72%|███████▏ | 449/625 [00:13<00:04, 40.21it/s]
- Sampling progress73%|███████▎ | 455/625 [00:13<00:03, 43.22it/s]
</pre>
- Sampling progress73%|███████▎ | 455/625 [00:13<00:03, 43.22it/s]
end{sphinxVerbatim}
Sampling progress : 73%|███████▎ | 455/625 [00:13<00:03, 43.22it/s]
- Sampling progress74%|███████▎ | 460/625 [00:13<00:03, 44.57it/s]
</pre>
- Sampling progress74%|███████▎ | 460/625 [00:13<00:03, 44.57it/s]
end{sphinxVerbatim}
Sampling progress : 74%|███████▎ | 460/625 [00:13<00:03, 44.57it/s]
- Sampling progress75%|███████▍ | 466/625 [00:13<00:03, 46.94it/s]
</pre>
- Sampling progress75%|███████▍ | 466/625 [00:13<00:03, 46.94it/s]
end{sphinxVerbatim}
Sampling progress : 75%|███████▍ | 466/625 [00:13<00:03, 46.94it/s]
- Sampling progress75%|███████▌ | 471/625 [00:13<00:03, 40.10it/s]
</pre>
- Sampling progress75%|███████▌ | 471/625 [00:13<00:03, 40.10it/s]
end{sphinxVerbatim}
Sampling progress : 75%|███████▌ | 471/625 [00:13<00:03, 40.10it/s]
- Sampling progress76%|███████▌ | 476/625 [00:13<00:03, 39.78it/s]
</pre>
- Sampling progress76%|███████▌ | 476/625 [00:13<00:03, 39.78it/s]
end{sphinxVerbatim}
Sampling progress : 76%|███████▌ | 476/625 [00:13<00:03, 39.78it/s]
- Sampling progress77%|███████▋ | 481/625 [00:14<00:03, 41.18it/s]
</pre>
- Sampling progress77%|███████▋ | 481/625 [00:14<00:03, 41.18it/s]
end{sphinxVerbatim}
Sampling progress : 77%|███████▋ | 481/625 [00:14<00:03, 41.18it/s]
- Sampling progress78%|███████▊ | 488/625 [00:14<00:02, 47.71it/s]
</pre>
- Sampling progress78%|███████▊ | 488/625 [00:14<00:02, 47.71it/s]
end{sphinxVerbatim}
Sampling progress : 78%|███████▊ | 488/625 [00:14<00:02, 47.71it/s]
- Sampling progress79%|███████▉ | 493/625 [00:14<00:02, 47.37it/s]
</pre>
- Sampling progress79%|███████▉ | 493/625 [00:14<00:02, 47.37it/s]
end{sphinxVerbatim}
Sampling progress : 79%|███████▉ | 493/625 [00:14<00:02, 47.37it/s]
- Sampling progress80%|███████▉ | 498/625 [00:14<00:02, 44.09it/s]
</pre>
- Sampling progress80%|███████▉ | 498/625 [00:14<00:02, 44.09it/s]
end{sphinxVerbatim}
Sampling progress : 80%|███████▉ | 498/625 [00:14<00:02, 44.09it/s]
- Sampling progress80%|████████ | 503/625 [00:14<00:03, 34.39it/s]
</pre>
- Sampling progress80%|████████ | 503/625 [00:14<00:03, 34.39it/s]
end{sphinxVerbatim}
Sampling progress : 80%|████████ | 503/625 [00:14<00:03, 34.39it/s]
- Sampling progress81%|████████▏ | 508/625 [00:14<00:03, 36.24it/s]
</pre>
- Sampling progress81%|████████▏ | 508/625 [00:14<00:03, 36.24it/s]
end{sphinxVerbatim}
Sampling progress : 81%|████████▏ | 508/625 [00:14<00:03, 36.24it/s]
- Sampling progress82%|████████▏ | 513/625 [00:14<00:02, 37.80it/s]
</pre>
- Sampling progress82%|████████▏ | 513/625 [00:14<00:02, 37.80it/s]
end{sphinxVerbatim}
Sampling progress : 82%|████████▏ | 513/625 [00:14<00:02, 37.80it/s]
- Sampling progress83%|████████▎ | 518/625 [00:15<00:02, 38.43it/s]
</pre>
- Sampling progress83%|████████▎ | 518/625 [00:15<00:02, 38.43it/s]
end{sphinxVerbatim}
Sampling progress : 83%|████████▎ | 518/625 [00:15<00:02, 38.43it/s]
- Sampling progress84%|████████▎ | 523/625 [00:15<00:02, 38.98it/s]
</pre>
- Sampling progress84%|████████▎ | 523/625 [00:15<00:02, 38.98it/s]
end{sphinxVerbatim}
Sampling progress : 84%|████████▎ | 523/625 [00:15<00:02, 38.98it/s]
- Sampling progress84%|████████▍ | 528/625 [00:15<00:02, 39.42it/s]
</pre>
- Sampling progress84%|████████▍ | 528/625 [00:15<00:02, 39.42it/s]
end{sphinxVerbatim}
Sampling progress : 84%|████████▍ | 528/625 [00:15<00:02, 39.42it/s]
- Sampling progress85%|████████▌ | 533/625 [00:15<00:02, 38.51it/s]
</pre>
- Sampling progress85%|████████▌ | 533/625 [00:15<00:02, 38.51it/s]
end{sphinxVerbatim}
Sampling progress : 85%|████████▌ | 533/625 [00:15<00:02, 38.51it/s]
- Sampling progress86%|████████▌ | 538/625 [00:15<00:02, 40.08it/s]
</pre>
- Sampling progress86%|████████▌ | 538/625 [00:15<00:02, 40.08it/s]
end{sphinxVerbatim}
Sampling progress : 86%|████████▌ | 538/625 [00:15<00:02, 40.08it/s]
- Sampling progress87%|████████▋ | 543/625 [00:15<00:01, 41.09it/s]
</pre>
- Sampling progress87%|████████▋ | 543/625 [00:15<00:01, 41.09it/s]
end{sphinxVerbatim}
Sampling progress : 87%|████████▋ | 543/625 [00:15<00:01, 41.09it/s]
- Sampling progress88%|████████▊ | 548/625 [00:15<00:01, 40.80it/s]
</pre>
- Sampling progress88%|████████▊ | 548/625 [00:15<00:01, 40.80it/s]
end{sphinxVerbatim}
Sampling progress : 88%|████████▊ | 548/625 [00:15<00:01, 40.80it/s]
- Sampling progress88%|████████▊ | 553/625 [00:15<00:01, 40.26it/s]
</pre>
- Sampling progress88%|████████▊ | 553/625 [00:15<00:01, 40.26it/s]
end{sphinxVerbatim}
Sampling progress : 88%|████████▊ | 553/625 [00:15<00:01, 40.26it/s]
- Sampling progress89%|████████▉ | 558/625 [00:16<00:01, 41.72it/s]
</pre>
- Sampling progress89%|████████▉ | 558/625 [00:16<00:01, 41.72it/s]
end{sphinxVerbatim}
Sampling progress : 89%|████████▉ | 558/625 [00:16<00:01, 41.72it/s]
- Sampling progress90%|█████████ | 563/625 [00:16<00:01, 43.32it/s]
</pre>
- Sampling progress90%|█████████ | 563/625 [00:16<00:01, 43.32it/s]
end{sphinxVerbatim}
Sampling progress : 90%|█████████ | 563/625 [00:16<00:01, 43.32it/s]
- Sampling progress91%|█████████ | 568/625 [00:16<00:01, 42.96it/s]
</pre>
- Sampling progress91%|█████████ | 568/625 [00:16<00:01, 42.96it/s]
end{sphinxVerbatim}
Sampling progress : 91%|█████████ | 568/625 [00:16<00:01, 42.96it/s]
- Sampling progress92%|█████████▏| 573/625 [00:16<00:01, 42.23it/s]
</pre>
- Sampling progress92%|█████████▏| 573/625 [00:16<00:01, 42.23it/s]
end{sphinxVerbatim}
Sampling progress : 92%|█████████▏| 573/625 [00:16<00:01, 42.23it/s]
- Sampling progress92%|█████████▏| 578/625 [00:16<00:01, 42.40it/s]
</pre>
- Sampling progress92%|█████████▏| 578/625 [00:16<00:01, 42.40it/s]
end{sphinxVerbatim}
Sampling progress : 92%|█████████▏| 578/625 [00:16<00:01, 42.40it/s]
- Sampling progress93%|█████████▎| 584/625 [00:16<00:00, 44.94it/s]
</pre>
- Sampling progress93%|█████████▎| 584/625 [00:16<00:00, 44.94it/s]
end{sphinxVerbatim}
Sampling progress : 93%|█████████▎| 584/625 [00:16<00:00, 44.94it/s]
- Sampling progress94%|█████████▍| 590/625 [00:16<00:00, 47.66it/s]
</pre>
- Sampling progress94%|█████████▍| 590/625 [00:16<00:00, 47.66it/s]
end{sphinxVerbatim}
Sampling progress : 94%|█████████▍| 590/625 [00:16<00:00, 47.66it/s]
- Sampling progress95%|█████████▌| 595/625 [00:16<00:00, 45.74it/s]
</pre>
- Sampling progress95%|█████████▌| 595/625 [00:16<00:00, 45.74it/s]
end{sphinxVerbatim}
Sampling progress : 95%|█████████▌| 595/625 [00:16<00:00, 45.74it/s]
- Sampling progress96%|█████████▌| 600/625 [00:16<00:00, 41.76it/s]
</pre>
- Sampling progress96%|█████████▌| 600/625 [00:16<00:00, 41.76it/s]
end{sphinxVerbatim}
Sampling progress : 96%|█████████▌| 600/625 [00:16<00:00, 41.76it/s]
- Sampling progress97%|█████████▋| 605/625 [00:17<00:00, 40.96it/s]
</pre>
- Sampling progress97%|█████████▋| 605/625 [00:17<00:00, 40.96it/s]
end{sphinxVerbatim}
Sampling progress : 97%|█████████▋| 605/625 [00:17<00:00, 40.96it/s]
- Sampling progress98%|█████████▊| 610/625 [00:17<00:00, 42.92it/s]
</pre>
- Sampling progress98%|█████████▊| 610/625 [00:17<00:00, 42.92it/s]
end{sphinxVerbatim}
Sampling progress : 98%|█████████▊| 610/625 [00:17<00:00, 42.92it/s]
- Sampling progress99%|█████████▊| 616/625 [00:17<00:00, 46.33it/s]
</pre>
- Sampling progress99%|█████████▊| 616/625 [00:17<00:00, 46.33it/s]
end{sphinxVerbatim}
Sampling progress : 99%|█████████▊| 616/625 [00:17<00:00, 46.33it/s]
- Sampling progress100%|█████████▉| 622/625 [00:17<00:00, 48.13it/s]
</pre>
- Sampling progress100%|█████████▉| 622/625 [00:17<00:00, 48.13it/s]
end{sphinxVerbatim}
Sampling progress : 100%|█████████▉| 622/625 [00:17<00:00, 48.13it/s]
- Sampling progress100%|██████████| 625/625 [00:17<00:00, 35.77it/s]
</pre>
- Sampling progress100%|██████████| 625/625 [00:17<00:00, 35.77it/s]
end{sphinxVerbatim}
Sampling progress : 100%|██████████| 625/625 [00:17<00:00, 35.77it/s]
22:19:39 INFO fit restored to maximum of posterior sampler_base.py:178
INFO fit restored to maximum of posterior sampler_base.py:178
Summary
-------
Number of Generations: 625
Number of Parameters: 2
Number of Walkers: 20
Number of Tuning Generations: 36
Scale Factor: 1.160638
Mean Integrated Autocorrelation Time: 3.2
Effective Sample Size: 3909.9
Number of Log Probability Evaluations: 66005
Effective Samples per Log Probability Evaluation: 0.059236
None
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | (9.91 +/- 0.16) x 10^-1 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (9.99 +/- 0.05) x 10^-2 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -6.598622 |
| total | -6.598622 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 17.903126 |
| BIC | 19.188708 |
| DIC | 17.120586 |
| PDIC | 1.961771 |
[9]:
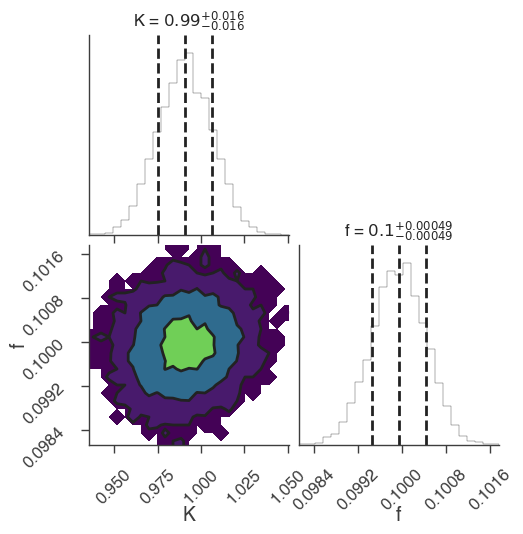
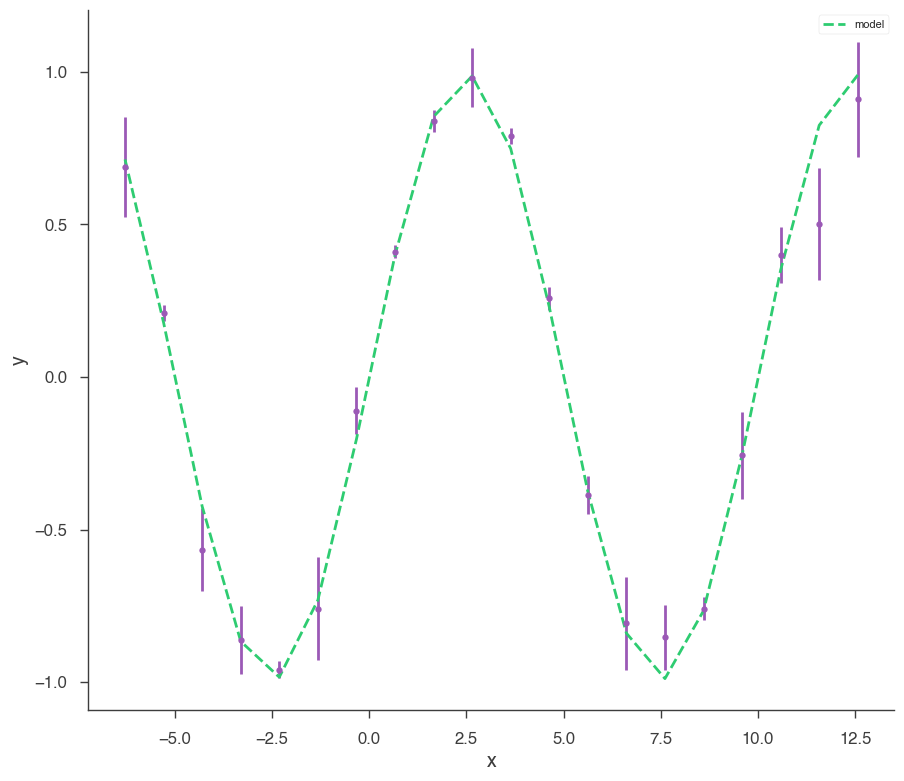
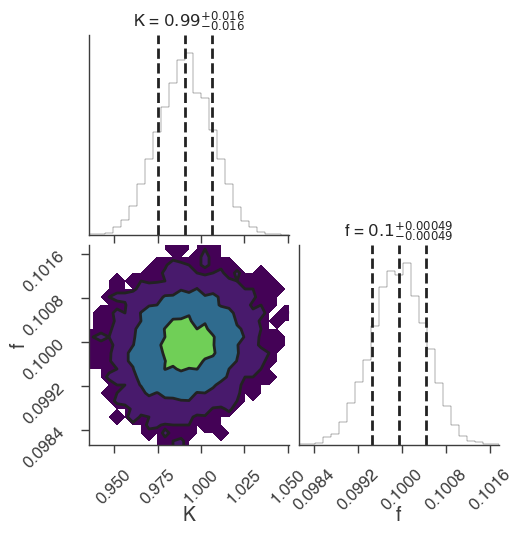
ultranest
[10]:
bayes_analysis.set_sampler("ultranest")
bayes_analysis.sampler.setup(
min_num_live_points=400, frac_remain=0.5, use_mlfriends=False
)
bayes_analysis.sample()
xyl.plot()
bayes_analysis.results.corner_plot()
22:19:40 INFO sampler set to ultranest bayesian_analysis.py:202
[ultranest] Sampling 400 live points from prior ...
Z=-inf(0.00%) | Like=-9686.44..-35.24 [-9686.4396..-3209.8402] | it/evals=0/401 eff=0.0000% N=400
Z=-6499.0(0.00%) | Like=-6479.79..-35.24 [-9686.4396..-3209.8402] | it/evals=40/442 eff=95.2381% N=400
Z=-5416.0(0.00%) | Like=-5405.93..-35.24 [-9686.4396..-3209.8402] | it/evals=79/489 eff=88.7640% N=400
Z=-5412.1(0.00%) | Like=-5363.98..-35.24 [-9686.4396..-3209.8402] | it/evals=80/490 eff=88.8889% N=400
Z=-5169.9(0.00%) | Like=-5161.98..-35.24 [-9686.4396..-3209.8402] | it/evals=90/502 eff=88.2353% N=400
Z=-4680.7(0.00%) | Like=-4662.57..-29.92 [-9686.4396..-3209.8402] | it/evals=120/533 eff=90.2256% N=400
Z=-4107.3(0.00%) | Like=-4099.40..-29.92 [-9686.4396..-3209.8402] | it/evals=160/588 eff=85.1064% N=400
Z=-3874.4(0.00%) | Like=-3861.87..-29.92 [-9686.4396..-3209.8402] | it/evals=180/617 eff=82.9493% N=400
Z=-3721.5(0.00%) | Like=-3702.89..-29.92 [-9686.4396..-3209.8402] | it/evals=200/642 eff=82.6446% N=400
Z=-3437.0(0.00%) | Like=-3417.06..-14.10 [-9686.4396..-3209.8402] | it/evals=240/699 eff=80.2676% N=400
Z=-3239.0(0.00%) | Like=-3231.59..-14.10 [-9686.4396..-3209.8402] | it/evals=270/745 eff=78.2609% N=400
Z=-3201.6(0.00%) | Like=-3189.07..-14.10 [-3198.8495..-2121.9750] | it/evals=280/761 eff=77.5623% N=400
Z=-2998.7(0.00%) | Like=-2989.38..-14.10 [-3198.8495..-2121.9750] | it/evals=320/831 eff=74.2459% N=400
Z=-2811.6(0.00%) | Like=-2804.86..-14.10 [-3198.8495..-2121.9750] | it/evals=360/925 eff=68.5714% N=400
Z=-2672.8(0.00%) | Like=-2664.04..-14.10 [-3198.8495..-2121.9750] | it/evals=395/996 eff=66.2752% N=400
Z=-2657.0(0.00%) | Like=-2647.70..-14.10 [-3198.8495..-2121.9750] | it/evals=400/1008 eff=65.7895% N=400
Z=-2518.7(0.00%) | Like=-2505.61..-14.10 [-3198.8495..-2121.9750] | it/evals=440/1109 eff=62.0592% N=400
Z=-2471.2(0.00%) | Like=-2463.31..-14.10 [-3198.8495..-2121.9750] | it/evals=450/1136 eff=61.1413% N=400
Z=-2388.4(0.00%) | Like=-2381.42..-14.10 [-3198.8495..-2121.9750] | it/evals=475/1198 eff=59.5238% N=400
Z=-2376.4(0.00%) | Like=-2354.43..-14.10 [-3198.8495..-2121.9750] | it/evals=480/1217 eff=58.7515% N=400
Z=-2239.2(0.00%) | Like=-2231.51..-14.10 [-3198.8495..-2121.9750] | it/evals=520/1306 eff=57.3951% N=400
Z=-2161.1(0.00%) | Like=-2149.53..-14.10 [-3198.8495..-2121.9750] | it/evals=556/1400 eff=55.6000% N=400
Z=-2143.9(0.00%) | Like=-2135.99..-14.10 [-3198.8495..-2121.9750] | it/evals=560/1408 eff=55.5556% N=400
Z=-2062.1(0.00%) | Like=-2055.04..-14.10 [-2121.2236..-1151.5067] | it/evals=600/1525 eff=53.3333% N=400
Z=-1994.7(0.00%) | Like=-1986.10..-14.10 [-2121.2236..-1151.5067] | it/evals=630/1633 eff=51.0949% N=400
Z=-1962.1(0.00%) | Like=-1951.90..-14.10 [-2121.2236..-1151.5067] | it/evals=640/1656 eff=50.9554% N=400
Z=-1935.2(0.00%) | Like=-1928.68..-14.10 [-2121.2236..-1151.5067] | it/evals=680/1735 eff=50.9363% N=400
Z=-1731.4(0.00%) | Like=-1720.45..-14.10 [-2121.2236..-1151.5067] | it/evals=720/1848 eff=49.7238% N=400
Z=-1508.3(0.00%) | Like=-1499.83..-14.10 [-2121.2236..-1151.5067] | it/evals=760/1916 eff=50.1319% N=400
Z=-1371.3(0.00%) | Like=-1357.55..-14.10 [-2121.2236..-1151.5067] | it/evals=800/1976 eff=50.7614% N=400
Z=-1331.4(0.00%) | Like=-1321.95..-14.10 [-2121.2236..-1151.5067] | it/evals=810/1990 eff=50.9434% N=400
Z=-1229.7(0.00%) | Like=-1220.04..-14.10 [-2121.2236..-1151.5067] | it/evals=840/2043 eff=51.1260% N=400
Z=-1059.6(0.00%) | Like=-1050.23..-14.10 [-1149.0606..-495.9028] | it/evals=880/2110 eff=51.4620% N=400
Z=-987.1(0.00%) | Like=-976.84..-14.10 [-1149.0606..-495.9028] | it/evals=900/2145 eff=51.5759% N=400
Z=-922.6(0.00%) | Like=-911.92..-14.10 [-1149.0606..-495.9028] | it/evals=920/2175 eff=51.8310% N=400
Z=-816.2(0.00%) | Like=-800.93..-14.10 [-1149.0606..-495.9028] | it/evals=960/2242 eff=52.1173% N=400
Z=-746.5(0.00%) | Like=-732.79..-14.10 [-1149.0606..-495.9028] | it/evals=990/2294 eff=52.2703% N=400
Z=-723.9(0.00%) | Like=-712.32..-14.10 [-1149.0606..-495.9028] | it/evals=1000/2307 eff=52.4384% N=400
Z=-651.9(0.00%) | Like=-642.68..-6.74 [-1149.0606..-495.9028] | it/evals=1040/2362 eff=53.0071% N=400
Z=-581.5(0.00%) | Like=-566.71..-6.74 [-1149.0606..-495.9028] | it/evals=1080/2431 eff=53.1758% N=400
Z=-513.5(0.00%) | Like=-504.23..-6.74 [-1149.0606..-495.9028] | it/evals=1120/2478 eff=53.8980% N=400
Z=-464.1(0.00%) | Like=-455.43..-6.74 [-495.8124..-274.9311] | it/evals=1160/2535 eff=54.3326% N=400
Z=-451.7(0.00%) | Like=-442.04..-6.74 [-495.8124..-274.9311] | it/evals=1170/2554 eff=54.3175% N=400
Z=-427.9(0.00%) | Like=-419.28..-6.74 [-495.8124..-274.9311] | it/evals=1200/2592 eff=54.7445% N=400
Z=-393.3(0.00%) | Like=-384.58..-6.74 [-495.8124..-274.9311] | it/evals=1240/2645 eff=55.2339% N=400
Z=-381.6(0.00%) | Like=-373.09..-6.74 [-495.8124..-274.9311] | it/evals=1260/2675 eff=55.3846% N=400
Z=-360.2(0.00%) | Like=-350.55..-6.74 [-495.8124..-274.9311] | it/evals=1280/2703 eff=55.5797% N=400
Z=-335.7(0.00%) | Like=-325.44..-6.74 [-495.8124..-274.9311] | it/evals=1320/2758 eff=55.9796% N=400
Z=-315.8(0.00%) | Like=-307.52..-6.74 [-495.8124..-274.9311] | it/evals=1350/2801 eff=56.2266% N=400
Z=-306.6(0.00%) | Like=-296.88..-6.74 [-495.8124..-274.9311] | it/evals=1360/2813 eff=56.3614% N=400
Z=-283.2(0.00%) | Like=-274.55..-6.74 [-274.6545..-160.8274] | it/evals=1400/2869 eff=56.7031% N=400
Z=-258.5(0.00%) | Like=-247.55..-6.74 [-274.6545..-160.8274] | it/evals=1440/2925 eff=57.0297% N=400
Z=-238.7(0.00%) | Like=-227.65..-6.74 [-274.6545..-160.8274] | it/evals=1480/2977 eff=57.4311% N=400
Z=-220.4(0.00%) | Like=-212.22..-6.74 [-274.6545..-160.8274] | it/evals=1520/3032 eff=57.7508% N=400
Z=-215.7(0.00%) | Like=-206.39..-6.74 [-274.6545..-160.8274] | it/evals=1530/3047 eff=57.8013% N=400
Z=-205.0(0.00%) | Like=-195.94..-6.74 [-274.6545..-160.8274] | it/evals=1560/3083 eff=58.1439% N=400
Z=-192.3(0.00%) | Like=-183.66..-6.74 [-274.6545..-160.8274] | it/evals=1600/3138 eff=58.4368% N=400
Z=-186.6(0.00%) | Like=-176.80..-6.74 [-274.6545..-160.8274] | it/evals=1620/3168 eff=58.5260% N=400
Z=-179.7(0.00%) | Like=-170.88..-6.74 [-274.6545..-160.8274] | it/evals=1640/3192 eff=58.7393% N=400
Z=-168.0(0.00%) | Like=-159.27..-6.74 [-160.7165..-88.0547] | it/evals=1680/3243 eff=59.0925% N=400
Z=-158.9(0.00%) | Like=-149.99..-6.74 [-160.7165..-88.0547] | it/evals=1710/3277 eff=59.4369% N=400
Z=-155.0(0.00%) | Like=-145.76..-6.74 [-160.7165..-88.0547] | it/evals=1720/3290 eff=59.5156% N=400
Z=-140.7(0.00%) | Like=-131.34..-6.74 [-160.7165..-88.0547] | it/evals=1760/3343 eff=59.8029% N=400
Z=-129.9(0.00%) | Like=-120.60..-6.74 [-160.7165..-88.0547] | it/evals=1800/3395 eff=60.1002% N=400
Z=-119.6(0.00%) | Like=-110.46..-6.74 [-160.7165..-88.0547] | it/evals=1840/3448 eff=60.3675% N=400
Z=-111.2(0.00%) | Like=-102.30..-6.74 [-160.7165..-88.0547] | it/evals=1880/3501 eff=60.6256% N=400
Z=-109.5(0.00%) | Like=-99.53..-6.74 [-160.7165..-88.0547] | it/evals=1890/3515 eff=60.6742% N=400
Z=-101.8(0.00%) | Like=-92.87..-6.74 [-160.7165..-88.0547] | it/evals=1920/3552 eff=60.9137% N=400
Z=-94.7(0.00%) | Like=-85.74..-6.74 [-88.0360..-48.8260] | it/evals=1960/3603 eff=61.1926% N=400
Z=-91.0(0.00%) | Like=-81.99..-6.74 [-88.0360..-48.8260] | it/evals=1980/3627 eff=61.3573% N=400
Z=-88.2(0.00%) | Like=-79.07..-6.74 [-88.0360..-48.8260] | it/evals=2000/3652 eff=61.5006% N=400
Z=-80.2(0.00%) | Like=-70.43..-6.74 [-88.0360..-48.8260] | it/evals=2040/3702 eff=61.7807% N=400
Z=-75.0(0.00%) | Like=-65.77..-6.74 [-88.0360..-48.8260] | it/evals=2070/3743 eff=61.9204% N=400
Z=-73.9(0.00%) | Like=-65.01..-6.74 [-88.0360..-48.8260] | it/evals=2080/3754 eff=62.0155% N=400
Z=-68.7(0.00%) | Like=-59.46..-6.74 [-88.0360..-48.8260] | it/evals=2120/3809 eff=62.1883% N=400
Z=-63.8(0.00%) | Like=-54.58..-6.74 [-88.0360..-48.8260] | it/evals=2160/3874 eff=62.1762% N=400
Z=-60.1(0.00%) | Like=-50.91..-6.74 [-88.0360..-48.8260] | it/evals=2200/3929 eff=62.3406% N=400
Z=-56.5(0.00%) | Like=-47.26..-6.74 [-48.8194..-28.4626] | it/evals=2240/3985 eff=62.4826% N=400
Z=-55.7(0.00%) | Like=-46.41..-6.74 [-48.8194..-28.4626] | it/evals=2250/4000 eff=62.5000% N=400
Z=-52.7(0.00%) | Like=-43.27..-6.74 [-48.8194..-28.4626] | it/evals=2280/4034 eff=62.7408% N=400
Z=-49.2(0.00%) | Like=-40.04..-6.74 [-48.8194..-28.4626] | it/evals=2320/4085 eff=62.9579% N=400
Z=-47.6(0.00%) | Like=-38.11..-6.74 [-48.8194..-28.4626] | it/evals=2340/4116 eff=62.9709% N=400
Z=-46.0(0.00%) | Like=-36.87..-6.74 [-48.8194..-28.4626] | it/evals=2360/4144 eff=63.0342% N=400
Z=-43.3(0.00%) | Like=-34.12..-6.74 [-48.8194..-28.4626] | it/evals=2400/4196 eff=63.2244% N=400
Z=-41.3(0.00%) | Like=-32.02..-6.74 [-48.8194..-28.4626] | it/evals=2430/4241 eff=63.2648% N=400
Z=-40.7(0.00%) | Like=-31.68..-6.74 [-48.8194..-28.4626] | it/evals=2440/4251 eff=63.3602% N=400
Z=-38.7(0.00%) | Like=-29.32..-6.65 [-48.8194..-28.4626] | it/evals=2480/4308 eff=63.4596% N=400
Z=-36.6(0.00%) | Like=-27.35..-6.65 [-28.4235..-18.4605] | it/evals=2520/4369 eff=63.4921% N=400
Z=-34.5(0.00%) | Like=-25.03..-6.65 [-28.4235..-18.4605] | it/evals=2560/4431 eff=63.5078% N=400
Z=-32.7(0.00%) | Like=-23.17..-6.65 [-28.4235..-18.4605] | it/evals=2600/4485 eff=63.6475% N=400
Z=-32.1(0.00%) | Like=-22.71..-6.65 [-28.4235..-18.4605] | it/evals=2610/4507 eff=63.5500% N=400
Z=-30.8(0.00%) | Like=-21.44..-6.65 [-28.4235..-18.4605] | it/evals=2640/4543 eff=63.7219% N=400
Z=-29.5(0.00%) | Like=-20.23..-6.65 [-28.4235..-18.4605] | it/evals=2680/4604 eff=63.7488% N=400
Z=-28.9(0.00%) | Like=-19.68..-6.65 [-28.4235..-18.4605] | it/evals=2700/4637 eff=63.7243% N=400
Z=-28.3(0.00%) | Like=-19.14..-6.65 [-28.4235..-18.4605] | it/evals=2720/4657 eff=63.8948% N=400
Z=-27.3(0.00%) | Like=-18.07..-6.65 [-18.4230..-14.4562] | it/evals=2760/4702 eff=64.1562% N=400
Z=-26.6(0.00%) | Like=-17.32..-6.65 [-18.4230..-14.4562] | it/evals=2790/4748 eff=64.1674% N=400
Z=-26.3(0.00%) | Like=-16.85..-6.65 [-18.4230..-14.4562] | it/evals=2800/4762 eff=64.1907% N=400
Z=-25.3(0.01%) | Like=-16.00..-6.65 [-18.4230..-14.4562] | it/evals=2840/4809 eff=64.4137% N=400
Z=-24.4(0.02%) | Like=-15.07..-6.65 [-18.4230..-14.4562] | it/evals=2880/4873 eff=64.3863% N=400
Z=-23.6(0.05%) | Like=-14.37..-6.65 [-14.4184..-13.9757] | it/evals=2920/4922 eff=64.5732% N=400
Z=-23.0(0.09%) | Like=-13.77..-6.65 [-13.7655..-13.7651]*| it/evals=2960/4977 eff=64.6712% N=400
Z=-22.8(0.11%) | Like=-13.57..-6.65 [-13.6204..-13.5711] | it/evals=2970/4990 eff=64.7059% N=400
Z=-22.4(0.17%) | Like=-13.06..-6.65 [-13.0593..-13.0500]*| it/evals=3000/5025 eff=64.8649% N=400
Z=-21.8(0.32%) | Like=-12.43..-6.65 [-12.4301..-12.4266]*| it/evals=3040/5077 eff=64.9989% N=400
Z=-21.5(0.42%) | Like=-12.22..-6.65 [-12.2217..-12.2213]*| it/evals=3060/5112 eff=64.9406% N=400
Z=-21.3(0.53%) | Like=-11.95..-6.65 [-11.9651..-11.9492] | it/evals=3080/5135 eff=65.0475% N=400
Z=-20.8(0.86%) | Like=-11.49..-6.65 [-11.4997..-11.4884] | it/evals=3120/5183 eff=65.2310% N=400
Z=-20.5(1.18%) | Like=-11.17..-6.63 [-11.1658..-11.1562]*| it/evals=3150/5222 eff=65.3256% N=400
Z=-20.4(1.31%) | Like=-11.04..-6.63 [-11.0442..-11.0422]*| it/evals=3160/5232 eff=65.3974% N=400
Z=-20.0(1.94%) | Like=-10.67..-6.63 [-10.6703..-10.6682]*| it/evals=3200/5279 eff=65.5872% N=400
Z=-19.6(2.77%) | Like=-10.26..-6.63 [-10.2578..-10.2452] | it/evals=3240/5327 eff=65.7601% N=400
Z=-19.3(3.87%) | Like=-9.82..-6.63 [-9.8347..-9.8185] | it/evals=3280/5382 eff=65.8370% N=400
Z=-19.0(5.26%) | Like=-9.55..-6.63 [-9.5525..-9.5512]*| it/evals=3320/5443 eff=65.8338% N=400
Z=-18.9(5.68%) | Like=-9.49..-6.63 [-9.4924..-9.4920]*| it/evals=3330/5454 eff=65.8884% N=400
Z=-18.7(7.06%) | Like=-9.30..-6.63 [-9.3151..-9.3049] | it/evals=3360/5487 eff=66.0507% N=400
Z=-18.4(9.10%) | Like=-9.05..-6.63 [-9.0473..-9.0322] | it/evals=3400/5540 eff=66.1479% N=400
Z=-18.3(10.32%) | Like=-8.90..-6.63 [-8.8996..-8.8970]*| it/evals=3420/5570 eff=66.1509% N=400
Z=-18.2(11.64%) | Like=-8.79..-6.63 [-8.8046..-8.7945] | it/evals=3440/5596 eff=66.2048% N=400
Z=-18.2(11.88%) | Like=-8.75..-6.63 [-8.7549..-8.7391] | it/evals=3446/5607 eff=66.1801% N=400
Z=-18.0(14.11%) | Like=-8.58..-6.61 [-8.5750..-8.5747]*| it/evals=3480/5663 eff=66.1220% N=400
Z=-17.8(16.14%) | Like=-8.43..-6.61 [-8.4316..-8.4249]*| it/evals=3510/5710 eff=66.1017% N=400
Z=-17.8(16.95%) | Like=-8.40..-6.61 [-8.4041..-8.3992]*| it/evals=3520/5724 eff=66.1157% N=400
Z=-17.6(20.12%) | Like=-8.25..-6.61 [-8.2492..-8.2477]*| it/evals=3560/5775 eff=66.2326% N=400
Z=-17.6(20.20%) | Like=-8.25..-6.61 [-8.2477..-8.2467]*| it/evals=3561/5776 eff=66.2388% N=400
Z=-17.5(23.33%) | Like=-8.11..-6.61 [-8.1083..-8.1057]*| it/evals=3600/5835 eff=66.2374% N=400
Z=-17.3(26.53%) | Like=-7.99..-6.61 [-7.9892..-7.9861]*| it/evals=3640/5889 eff=66.3144% N=400
Z=-17.2(29.84%) | Like=-7.83..-6.61 [-7.8296..-7.8294]*| it/evals=3680/5943 eff=66.3900% N=400
Z=-17.2(30.78%) | Like=-7.81..-6.61 [-7.8121..-7.8086]*| it/evals=3690/5960 eff=66.3669% N=400
Z=-17.1(33.39%) | Like=-7.74..-6.61 [-7.7368..-7.7321]*| it/evals=3720/5993 eff=66.5117% N=400
Z=-17.0(36.81%) | Like=-7.61..-6.61 [-7.6118..-7.6098]*| it/evals=3760/6041 eff=66.6548% N=400
Z=-16.9(38.67%) | Like=-7.55..-6.61 [-7.5541..-7.5501]*| it/evals=3780/6074 eff=66.6197% N=400
Z=-16.9(40.50%) | Like=-7.50..-6.61 [-7.5000..-7.4996]*| it/evals=3800/6097 eff=66.7018% N=400
Z=-16.8(43.96%) | Like=-7.41..-6.61 [-7.4130..-7.4117]*| it/evals=3840/6152 eff=66.7594% N=400
Z=-16.8(46.62%) | Like=-7.36..-6.61 [-7.3563..-7.3562]*| it/evals=3870/6199 eff=66.7356% N=400
Z=-16.7(47.44%) | Like=-7.33..-6.61 [-7.3341..-7.3316]*| it/evals=3880/6211 eff=66.7699% N=400
[ultranest] Explored until L=-7
[ultranest] Likelihood function evaluations: 6243
[ultranest] logZ = -15.99 +- 0.09775
[ultranest] Effective samples strategy satisfied (ESS = 983.7, need >400)
[ultranest] Posterior uncertainty strategy is satisfied (KL: 0.45+-0.05 nat, need <0.50 nat)
[ultranest] Evidency uncertainty strategy is satisfied (dlogz=0.42, need <0.5)
[ultranest] logZ error budget: single: 0.14 bs:0.10 tail:0.41 total:0.42 required:<0.50
[ultranest] done iterating.
22:19:52 INFO fit restored to maximum of posterior sampler_base.py:178
INFO fit restored to maximum of posterior sampler_base.py:178
Maximum a posteriori probability (MAP) point:
| result | unit | |
|---|---|---|
| parameter | ||
| demo.spectrum.main.Sin.K | (9.92 -0.16 +0.15) x 10^-1 | 1 / (cm2 keV s) |
| demo.spectrum.main.Sin.f | (9.99 +/- 0.05) x 10^-2 | rad / keV |
Values of -log(posterior) at the minimum:
| -log(posterior) | |
|---|---|
| demo | -6.602016 |
| total | -6.602016 |
Values of statistical measures:
| statistical measures | |
|---|---|
| AIC | 17.909915 |
| BIC | 19.195497 |
| DIC | 17.190724 |
| PDIC | 1.996892 |
| log(Z) | -6.946315 |
[10]: